Figure 4.
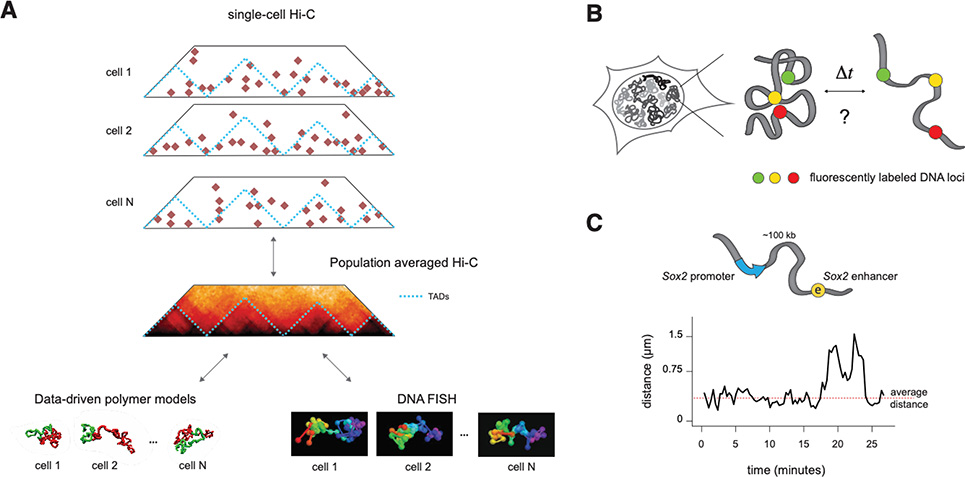
A. Single-cell Hi-C (top), polymer simulations (bottom left) and super-resolution DNA FISH (bottom right) show that population-averaged signals in 3C methods arise from highly variable structures that occur simultaneously in single cells.
B. To measure the temporal dynamics of chromosome structures, experiments are needed where the spatial positions of two or more chromosomal locations in cis can be measured in time, in living cells.
C. Simultaneous live-cell imaging of genomic locations flanking the Sox2 promoter and its (super-) enhancer in mESC shows that their distances fluctuate around average values over an experimental timescale of two hours, with sporadic stochastic larger changes. Adapted from. (Alexander et al. 2019).