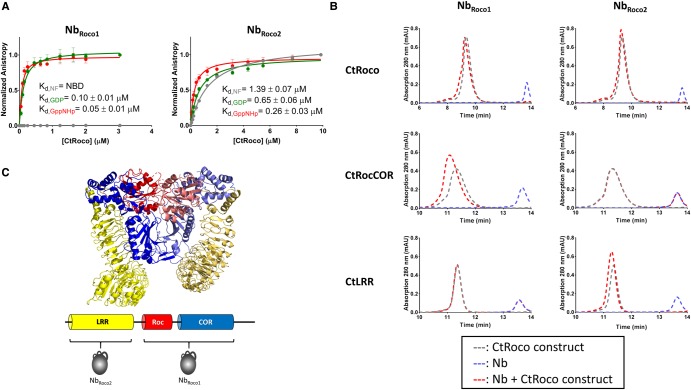
Figure 1. NbRoco1 and NbRoco2 are conformation-specific Nbs targeting different domains of CtRoco.
(A) Influence of the nucleotide-bound state of CtRoco on the affinity for NbRoco1 (left) and NbRoco2 (right) assessed by fluorescence anisotropy titrations. The fluorescence anisotropy signal of the FITC-labeled Nbs is monitored upon titration with increasing concentrations of CtRoco in the nucleotide-free state (gray), or bound to GDP (green) or GppNHp (red). The corresponding equilibrium dissociation constants (Kd ± standard error) obtained by fitting with a quadratic binding equation are given (each data point is the average of three independent measurements with the error bars representing the standard deviation; NBD = no binding detectable at the concentrations used). (B) Analysis of the domain specificity of NbRoco1 (left) and NbRoco2 (right) using analytical size exclusion chromatography. NbRoco1 and NbRoco2 were mixed with a small molar excess of the CtRoco protein or either of its constituting domains CtRocCOR or CtLRR, and samples were analyzed on analytical size exclusion chromatography (chromatograms shown as red dotted lines). These elution profiles are compared with the elution profiles of either the Nbs or the CtRoco constructs individually (blue and gray dotted lines, respectively). Binding of the Nbs to CtRoco and to either of its constituting domains, is reflected by a shift of the elution peak corresponding to the CtRoco construct (red vs. gray curve) and a disappearance of the peak corresponding to the Nb elution volume (blue curve). (C) Cartoon representation of the X-ray crystal structure of the homodimer of the LRR-RocCOR construct of CtRoco with the LRR domains shown in yellow, the Roc domains in red and the COR domains in blue. Corresponding domains from adjacent subunits in the homodimer are shown in different shades (PDB code 6HLU) [37]. Below, the domain arrangement of a CtRoco subunit is shown schematically with the binding epitopes of NbRoco1 and NbRoco2 indicated.