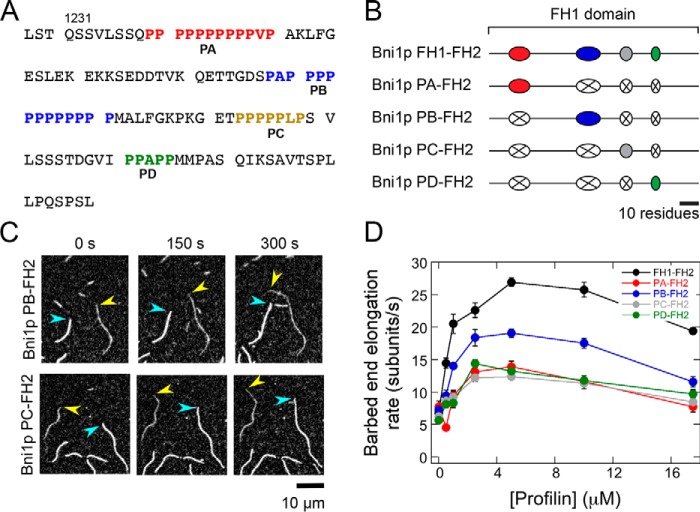
Figure 2.
Individual polyproline tracts robustly mediate actin polymerization by Bni1p. The experimental conditions were as follows: 0.75 μm actin (33% Oregon Green–labeled) in microscopy buffer with varying concentrations of S. cerevisiae profilin. The data were collected by TIRF microscopy. A, FH1 domain sequence, which consists of amino acids 1228–1347 of Bni1p. The four polyproline tracts are color-coded in red (PA), blue (PB), gold (PC), and green (PD). B, schematic representations of the FH1 domain of WT and single-tract variant Bni1p constructs. Red, blue, gray, and green ovals represent polyproline tracts PA, PB, PC, and PD, and black lines represent nonproline sequences. In each Bni1p single-tract variant, three tracts have been substituted with poly(Gly/Ser) sequences that correspond to the length of each replaced tract (represented by a white oval with an X). C, representative series of TIRF micrographs of elongating actin filaments in the presence of single-tract Bni1p variants and 5 μm S. cerevisiae profilin. Micrographs were collected at 150-s intervals. Yellow arrowheads indicate barbed ends of filaments bound by a Bni1p construct. Blue arrowheads indicate barbed ends of control (i.e. not formin-bound) filaments. D, dependence of the actin filament elongation rates mediated by Bni1p FH1FH2 (black circles), Bni1p PA–FH2 (red circles), Bni1p PB–FH2 (blue circles), Bni1p PC-FH2 (gray circles), and Bni1p PD–FH2 (green circles) on the concentration of profilin. Error bars are the standard errors of the mean elongation rates of at least 10 filaments.