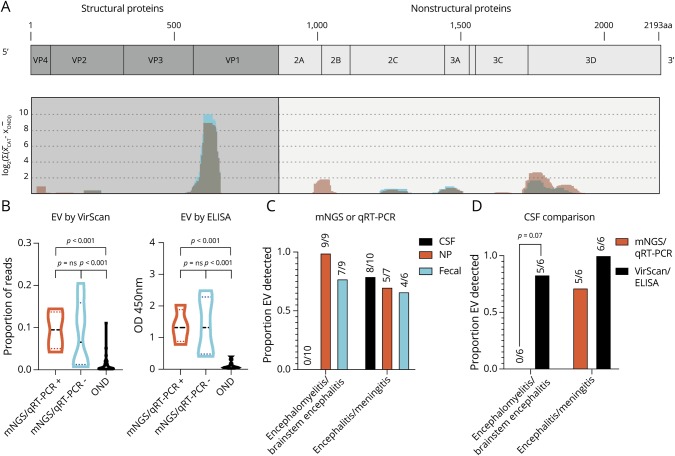
Figure 3. VirScan Identifies immunodominant enterovirus antigens and improves the detection of CSF enterovirus in encephalomyelitis/brainstem encephalitis.
(A) VirScan identified 136 unique, enriched viral antigens with taxonomies linking them to genus Enterovirus. We mapped 123 of these 136 peptides with BLASTP to a reference EV-A71 genome (Genbank Accession AXK59213.1), as described previously9 (coding genes in light blue, non-coding genes in orange). Mapping revealed the relative locations of the EV antigens identified in the Catalonia cases (graphed blue shading) across the EV genome, which appeared remarkably conserved, as seen previously in pediatric acute flaccid myelitis (graphed light red shading with overlap appearing gray).10 (B) Violin plot revealing enrichment for EV antigens by VirScan and EV VP1 antigen by ELISA in Catalonia cases, regardless of whether an EV had been previously identified by mNGS or qRT-PCR (p = ns for both comparisons). In both groups, EV detection was significantly greater than in the pediatric OND controls (p < 0.001 for all comparisons as indicated). The Mann-Whitney test was used, with Bonferroni correction for VirScan. (C) Differences in virus detection for subjects with encephalomyelitis/brainstem encephalitis or encephalitis/meningitis. The detection of CSF EV by mNGS or qRT-PCR was low (0/10). (D) VirScan or ELISA improved EV detection in encephalomyelitis/brainstem encephalitis (0/6 vs 5/6, p = 0.07 by the McNemar test). CSF = cerebral spinal fluid; EV = enterovirus; mNGS = metagenomic next-generation sequencing; NP = nasopharyngeal; OND = other neurologic disease.