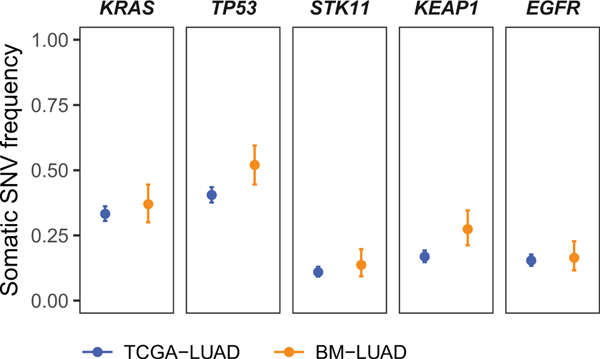
Extended Data Fig. 3.
Power analysis and statistical simulation of case-control study.
Single nucleotide variants (SNVs) and short insertions/deletions (indels) in BM-LUAD were analyzed by MutSig2CV and dNdScv to identify driver genes under positive selection. Identified drivers are statistically significant by both MutSig2CV and dNdScv at 1% false discovery rate, except for EGFR, which harbors recurrent indels that are considered only by MutSig2CV. The mutation frequencies of the identified drivers are shown for BM-LUAD and TCGA-LUAD after matching adjustment by coarsened exact matching, and statistical significances of differences in mutation frequency were assessed by weighted logistic regression using the matching weights. None of the identified drivers were statistically significantly different between BM-LUAD and TCGA-LUAD at 0.05 significance level with Benjamini-Hochberg multiple hypothesis correction.