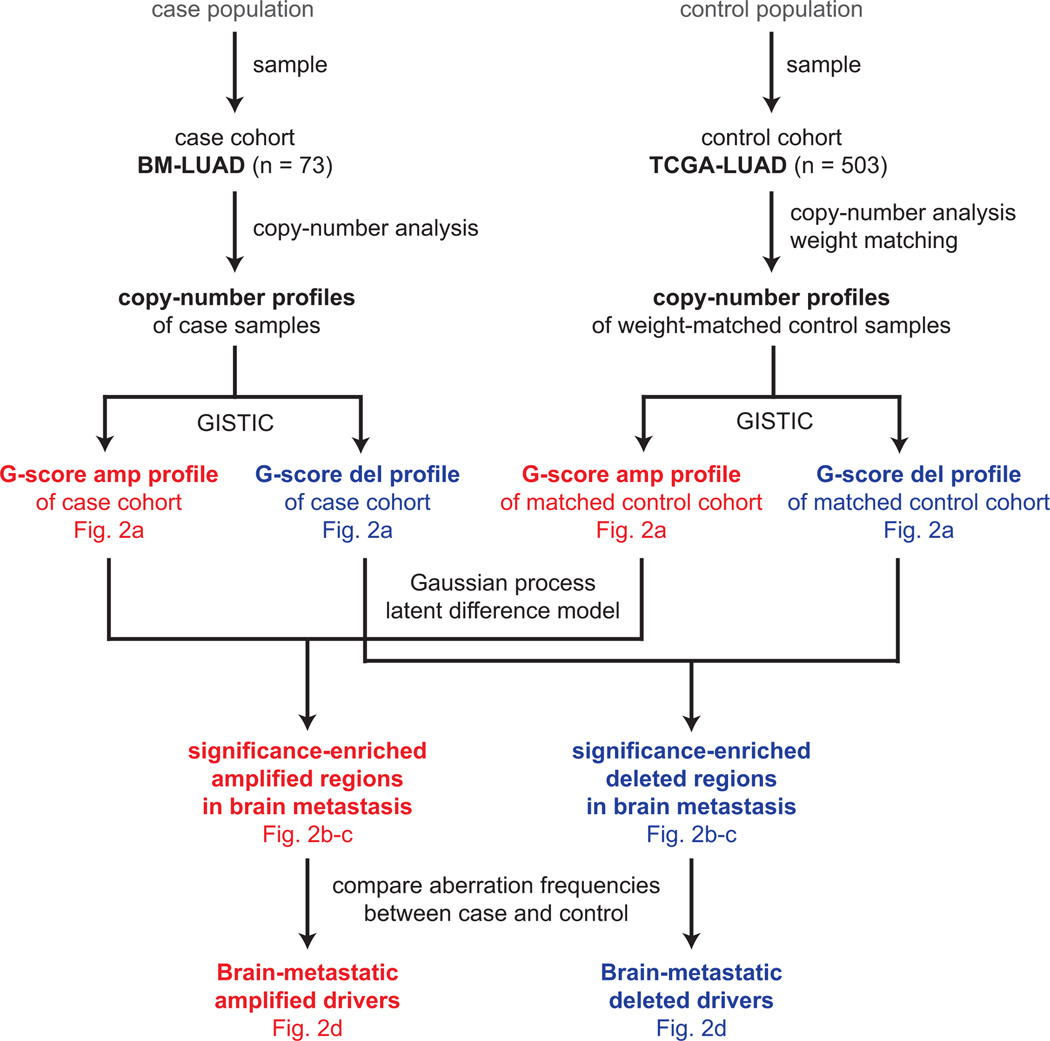
Extended Data Fig. 5.
Power analysis and statistical simulation of case-control study.
CNAs Somatic copy-number profiles in case cohort (BM-LUAD) and weight-matched control cohort (TCGA-LUAD) were analyzed by GISTIC. Copy-number profiles of control samples were multiplied by matching weights, which were defined to balance covariate distributions between case and control cohorts using the coarsened exact matching method. G-score profiles for amplifications and deletions were independently analyzed by a Gaussian process latent difference model to identify significantly enriched regions. Candidate drivers were identified by logistic regression comparing aberration frequencies between case and weighted controls; the candidates were further validated in an independent cohort by fluorescence in situ hybridization.