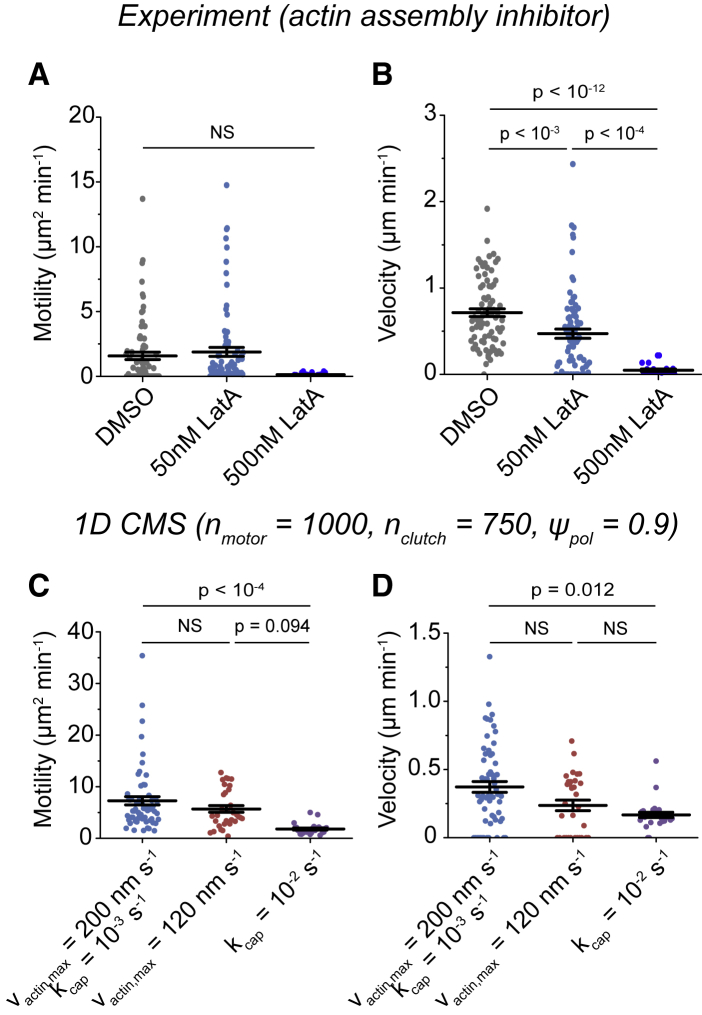
Figure 6.
Simulated and experimental predictions of the effects of actin polymerization inhibitors on confined glioma cell migration. (A) The motility coefficients for U251 cells expressing eGFP-β-actin and treated with vehicle control (DMSO) or 50 or 500 nM LatA are shown (n = 78, 81, 23 cells). (B) The velocities for the experimental conditions in (A) are shown. (C) The motility coefficients from simulations using a reference parameter set (Table S1; vactin,max = 200 nm s−1, kcap = 0.001 s−1) or simulations in which vactin,max was reduced (vactin,max = 120 nm s−1) or in which kcap was increased (kcap = 0.01 s−1) are shown (n = 60, 28, and 24 simulations). All simulations had nmotor = 1000, nclutch = 750, and ψpol = 0.9, and all other parameter values are reported in Table S1. (D) The velocities from the simulations in (C) are shown. Individual motility coefficients and velocities were obtained from fits to Eq. 2. The error bars represent mean ± SEM, NS denotes no significant difference, and p > 0.01 by one-way Kruskal-Wallis ANOVA. To see this figure in color, go online.