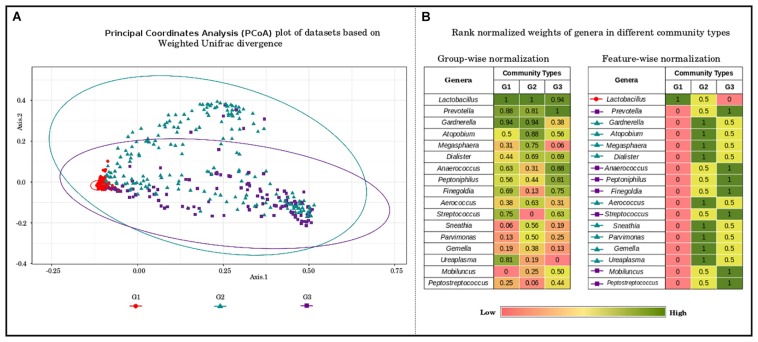
FIGURE 3.
(A) Principal coordinate analysis (PcoA) plot of datasets based on Weighted Unifrac divergence, representing three distinct community types. The samples analyzed clustered into three distinct clusters/community types based on Weighted Unifrac divergence as a distance metric. Dirichlet Multinomial Mixtures probabilistic model was employed to obtain statistically optimum number of clusters/community types. The corresponding Dirichlet model fit plot representing the number of optimum clusters/community types for the datasets analyzed is provided as Supplementary Figure S2. (B) Rank normalized weights of genera in different community types. The genera constituting the vaginal microbial communities in the datasets analyzed were ranked based on their weights/probabilities in (a) each community type/cluster and (b) throughout different community types/clusters. This exercise was performed to identify the driving/contributing bacterial taxa/groups for the observed clusters. The community type G1, constituting most of the samples from the stages analyzed is observed to be driven solely by members of Lactobacilli. The genera namely, Gardnerella, Atopobium, Megasphaera, Dialister, Aerococcus, Sneathia, Parvimonas, Gemella, and Ureaplasma are seen to drive/constitute the community type G2. The driving taxa for community type G3 are observed to be Prevotella, Anaerococcus, Peptoniphilus, Finegoldia, Streptococcus, Mobiluncus, and Peptostreptococcus.