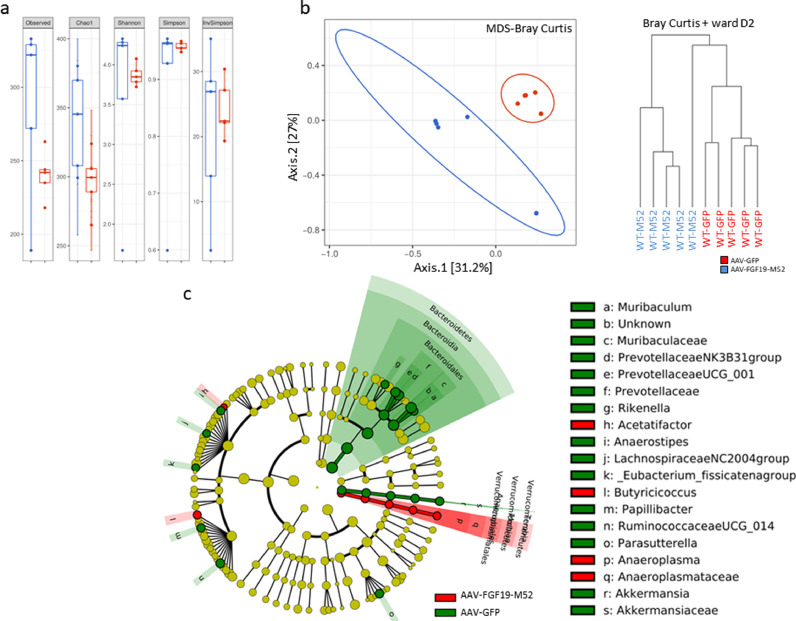
Fig. 3.
AAV-FGF19-M52 increases alpha diversity and generate a beneficial shift in the gut microflora of WT mice. (a) Graphs represent the alpha diversity at the OTU level in WT and Fxrnull mice. Observed and Chao1 indexes calculate the alpha diversity in term of richness (number of taxa that are present in the samples). Shannon, Simpson and inversed Simpson indexes calculate the alpha diversity regarding the evenness of taxa in the samples. (b) Graphs represent the distance between samples using the OTU distribution of each sample. The distance is here represented on 2 axes summarizing the entire distribution of all the OTU present in the samples (MDS representation) and a hierarchical clustering tree (dendogram). (c) Visual comparison of the relative abundance barplots indicates differences between the groups as also seen in the pairwise group LEfSe analyses (log(LDA Score)>2.0). The cladograms represented here indicate the bacterial taxa that are significantly different between the 2 groups being compared. This analysis helps to identify a first selection of differential bacterial taxa in the considered groups. (Overlapping taxa on the green section: Verrucomicrobia, Verrucomicrobiae, Verrucomicrobiales. Overlapping taxa on the red section:Tenericutes, Mollicutes, Anaeroplasmatales)