Figure 2.
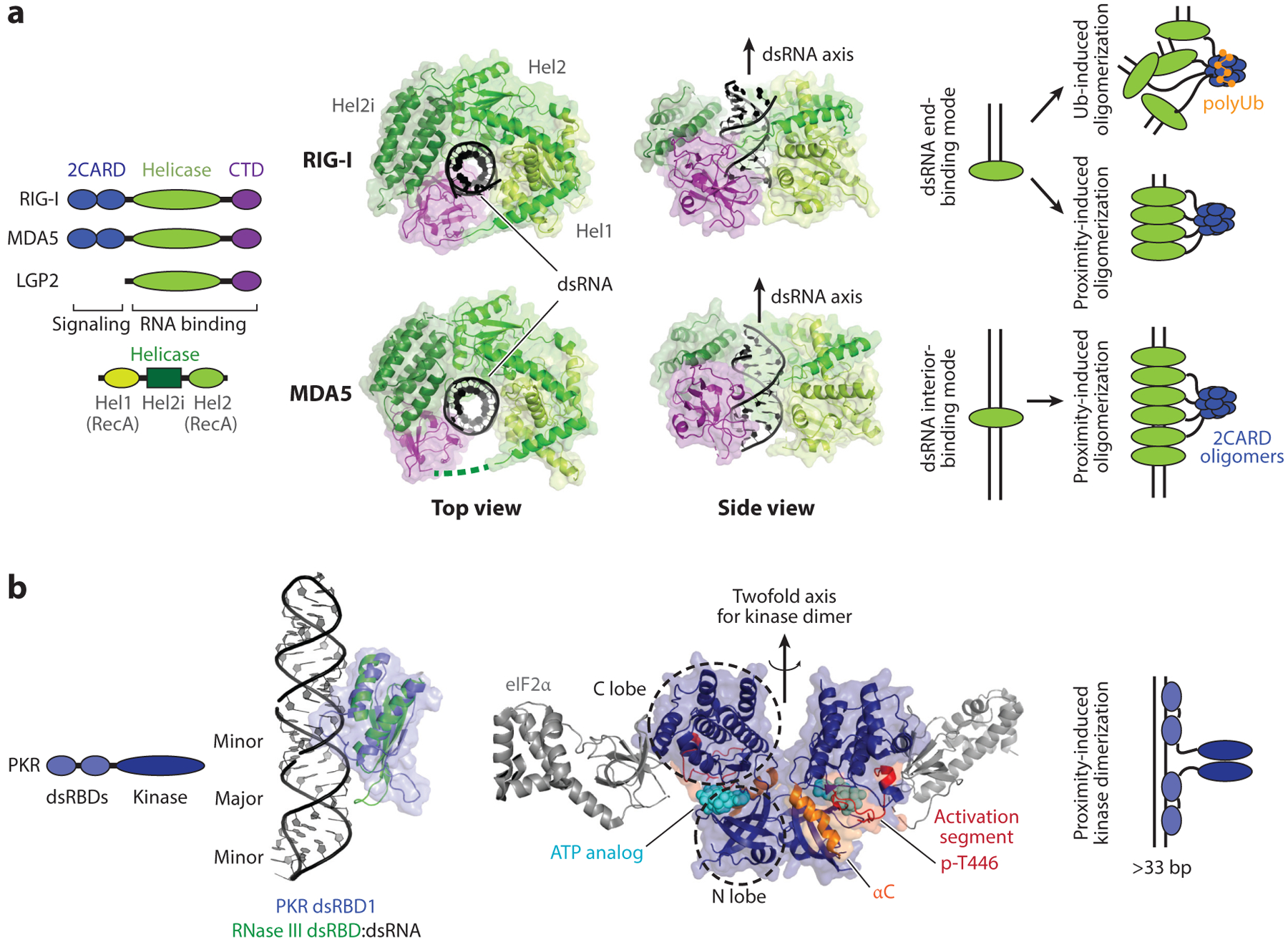
RLR and PKR. (a) Domain architectures and structures of human RIG-I and MDA5 in complex with dsRNA (PDB: 3TMI and 4GL2). RIG-I binds the dsRNA end as a monomer (as shown in the crystal structure), but on long dsRNA, it assembles into filamentous oligomers. Filament formation of RIG-I on dsRNA promotes 2CARD tetramerization through a proximity-induced mechanism. The K63-linked polyubiquitin chain also promotes 2CARD tetramerization. In contrast to RIG-I, MDA5 only binds the dsRNA interior, not the end, and its filament formation is obligatory. As with RIG-I, 2CARD tetramerization (with or without the K63-linked polyubiquitin chain) is required for antiviral signal activation. (b) Domain architecture and structures of PKR. The structure of PKR dsRBDs in complex with dsRNA is not available, but studies suggest that PKR dsRBDs bind dsRNA in a manner similar to other canonical dsRBDs. The RNA-free structure of PKR dsRBD1 (blue) (PDB: 1QU6) is overlaid on the structure of Aquifex aeolicus RNase III in complex with dsRNA (green) (PDB: 2EZ6). On the right is the structure of the PKR kinase domain (dark blue) in the active dimeric state and in complex with eIF2α (gray) (PDB: 2A19). Analogous to RLRs, PKR utilizes dsRNA binding and the proximity-induced mechanism to dimerize and activate the kinase domain. Abbreviations: CTD, C-terminal domain; dsRBD, dsRNA-binding domain; PKR, protein kinase R; RLR, RIG-I-like receptor; Ub, ubiquitin.
