Abstract
Accumulating evidences suggest that miRNAs may prove essential therapeutic targets for the treatment of cancer. Herein, the role and therapeutic implications of miRNA-143 was investigated in gastric cancer. The results revealed miRNA-143 to be aberrantly downregulated in gastric cancer cell lines. Ectopic expression of miRNA-143 resulted in a significant (P<0.05) inhibition of AGS gastric cancer cell proliferation suggestive of the tumor suppressive role of miRNA-143. The inhibition of AGS cell proliferation was mainly via activation of apoptotic cell death as evident from the AO/EB and annexin V/PI staining. Additionally, miR-143 overexpression also caused a significant (P<0.05) decline in the migration and invasion of AGS cells. TargetScan analysis and the dual luciferase assay revealed STAT3 to be the potential target of miRNA-143 in AGS cells. Analysis of STAT3 expression in gastric cancer cell lines showed upto 3.5 fold upregulation of STAT3. However, miRNA-143 overexpression resulted in considerable downregulation of STAT3 expression. Silencing of STAT3 also resulted in the inhibition of cell proliferation, migration and invasion of the AGS cells. Moreover, overexpression of STAT3 could nullify the tumor suppressive effects of miRNA-143 in AGS cells. Taken together, miRNA-143 has a tumor suppressive role in gastric cancer and may prove essential in gastric cancer treatment.
Keywords: Gastric cancer, tumor suppressor, miRNA-143, migration, apoptosis
Introduction
MicroRNAs (miRNAs) include RNA molecules that are around 23 nucleotides in length and regulate the expression of a number of genes in human and other organisms. The miRs bind to the messenger RNAs (mRNAs) and enforce their post transcriptional repression [1]. The miRNAs modulate the expression of about 30% of the human protein coding genes. They are involved in different cellular and physiological processes such as cell cycle, proliferation, and apoptosis to name a new [2]. Accumulating evidences suggest that microRNAs are aberrantly expressed in cancer cells and are therefore considered to be prospective therapeutic targets/agents for the treatment of cancer [3]. The miRNA-143-3p has been shown to be involved in the proliferation and metastasis of many cancers [4]. It has been reported to act as tumor suppressor in colon and breast cancer [5,6]. Similarly, it has been shown to regulate the proliferation and invasion of breast cancer cells by targeting MAPK7 [7]. Although a few studies report the involvement of miR-143 in gastric cancer, the role of miR-143 in gastric cancer is yet to be investigated thoroughly. Gastric cancer is one of the commonly detected cancers and accounts for significant human mortality. In 2002, gastric cancer caused 0.65 million deaths and 0.9 million new cases of gastric cancer were detected [8,9]. It has been reported that gastric cancer incidence varies with geography. Nonetheless, developing countries have comparatively higher incidences of gastric cancer [10,11]. Although recent reports have shown considerable improvement in the 5-year survival of gastric, but the survival rates are still lower than in case of other cancers [12]. Dearth of efficient chemotherapeutic drugs and therapeutic targets imposes hurdles in gastric cancer treatment [13]. Herein, we report the role of miR-143 as tumor suppressor in gastric cancer. It was observed that miR-143 is aberrantly downregulated in gastric cancer and restoration of miR-143 expression suppresses the growth, migration and invasion of the gastric cancer cells by suppressing STAT3 expression. Taken together, the present study indicates that miR-143 may prove essential in the treatment of Gastric cancer.
Materials and methods
Cell lines and culture conditions
Gastric cancer cell lines (AGS, SNU-1, SNU-5, SNU-16, NCIN87 and KATOIII) were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) containing penicillin (100 U/mL), streptomycin (100 μg/ml) (Sigma-Aldrich, St. Louis, MO, USA), and 10% fetal bovine serum (FBS; Gibco) at 37°C in 5% CO2.
cDNA synthesis and quantitative RT-PCR
The TRIzol reagent (Invitrogen) was used for the extraction of RNA from all the cell lines. This was followed by purification of the RNA by RNeasy Mini Kit (Qiagen). The complementary DNA was then synthesized with the help of miScript Reverse Transcription Kit (Qiagen). Afterwards, the cDNA was amplified by using SYBR Premix Ex Taq™ (TaKaRa, Otsu, Shiga, Japan). The expression was estimated by 2-ΔΔCt method as described previously [14]. Actin was used as an internal control.
Cell transfection
The miR-143 mimics and NC were synthesized by RiboBio (Guangzhou, China). The transfection was then carried out by the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s instructions. As the AGS cells reached 80% confluence, the appropriate concentrations of miR-143 mimics or NC were transfected into AGS cells.
Cell counting kit-8 (CCK-8) assay
After 48 h of transfection, the AGS cells were grown in 96-well plates in three replicates at the density of 1.0×103 cells/well. The proliferation rate and the viability of the AGS cells were determined by Cell Counting Kit-8 following the guidelines of the manufacturer. Around 10 µL CCK-8 reagent was added into the wells at specific time intervals (12, 24, 48, 72 and 96 h) and the plates were then subjected to incubation for about 2 h at 37°C. The cell viability was then measured by taking the absorbance at 450 nm.
Apoptosis assays
For AO/EB staining, the transfected gastric cancer AGS cells (0.6×106) were grown in 6 well plates. Following an incubation period of around 12 hours, the AGS cells were subjected to incubation for 24 h at 37°C. As the cells sloughed off, 25 μl cell cultures were put onto a glass slides and subjected to staining with a solution (1 μl) of AO and EB. The slides were covered with a covers lip and examined with a fluorescent microscope. Annexin V/PI staining of the AGS cells was performed as described previously [15].
Target identification and dual luciferase assay
The miR-143 target was identified by TargetScan online software (http://www.targetscan.org). The miR-143 mimics or NC were co-transfected with Plasmid pGL3-STAT3-3’-UTR-WT or pGL3-STAT3-3’-UTR-MUT into AGS cells. Dual-luciferase reporter assay (Promega) was carried out at 48 h post-transfection. Renilla was luciferase used for normalization.
Cell migration and Invasion assay
The gastric AGS cancer cells were subjected to transfection with miR-143 mimics and cultured for 12 h. The transwell assays were then used for determining the effects of miR-143 overexpression on AGS cells. In brief, 250 µl transfected AGS cell suspension at the density of 4×105/ml were placed in upper chamber while as only media was placed on the lower chamber. This followed by incubation for 24 h. The cells that moved to the lower chamber were fixed using glacial acetic acid. The cells were subsequently stained with crystal violet for 30 min and observed under a microscope. In case of cell invasion assay, Martigel was used and rest of the procedure was same.
Western blotting
The AGS cells were firstly subjected to washing with ice-cold PBS and the suspended in a lysis buffer at 4°C and then shifted to 95°C. Thereafter, the protein content of each cell extract was checked by Bradford assay. About, 40 µg of protein was loaded from each sample and separated by SDS-PAGE before being shifted to polyvinylidene fluoride membrane. The membranes were then subjected to treatment with TBS and then exposed to primary antibodies at 4°C. Thereafter, the cells were treated with appropriate secondery antibodies and the proteins of interest were visualised by enhanced chemiluminescence reagent.
Statistical analysis
Data are shown as mean ± SD. Statistical analysis was done using Students t-test with GraphPad prism 7 software. Values of P<0.05 were taken as indicative of significant difference.
Results
miRNA-143 is aberrantly downregulated in gastric cancer cell lines
The expression profile of miRNA-143 was investigated in normal and gastric cancer cell lines by qRT-PCR (Figure 1A). It was found that miRNA-143 exhibited aberrant expression and was significantly (P<0.05) downregulated in all the gastric cancer cell lines. It was revealed that miRNA-143 is downregulated in gastric cancer lines by upto 6 fold relative to the normal cell line. The highest downregulation was found in case of the AGS cell line.
Figure 1.
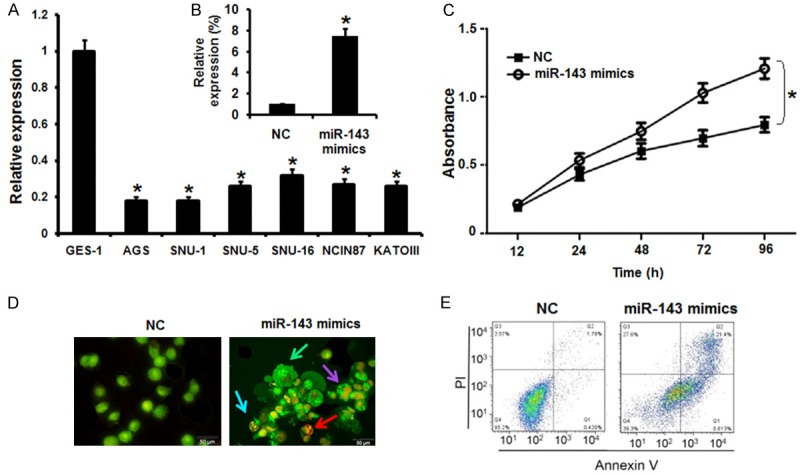
Overexpression of miR-143 inhibits the proliferation of gastric cancer cells. A. Expression of miR-143 in GES-1 normal and six different lung cancer cell lines. B. Expression of miR-143 in NC or miR-143 mimics transfected AGS gastric cancer cells. C. CCK-8 assay showing the % viability of NC or miR-143 mimics transfected AGS gastric cancer cells at indicated time intervals. D. AO/EB staining showing induction of apoptosis in NC or miR-143 mimics transfected AGS gastric cancer cells. E. Annexin V/PI staining showing percentage of apoptosis in NC or miR-143 mimics transfected AGS gastric cancer cells. The experiments were performed in triplicate and expressed as mean ± SD (*P<0.05).
Tumor suppressive role of miR-143 in gastric cancer
Next, we sought to understand the role of miRNA-143 in gastric cancer and for that the AGS cells were transfected with either the miRNA-143 mimics or NC (negative control). The overexpression of miRNA-143 in AGS cells was confirmed by the qRT-PCR. It was revealed that miRNA-143 overexpression caused around 7.5 fold increase in the expression of miRNA-143 relative to NC transfected cells (Figure 1B). Next the proliferation rate of the NC and miRNA-143 mimics transfected AGS cells was determined by CCK-8 assay and it was found that the miRNA-143 overexpression caused significant decline in the proliferation rate of the AGS cells (Figure 1C). Next, AO/EB and annexin V/PI staining of the NC and miRNA-143 transfected cells were performed to unveil the underlying mechanism for the decrease in the proliferation rate of the miR-143 mimics transfected cells. The AO/EB staining results showed that miRNA-143 overexpression activated apoptotic cell death of the AGS cells (Figure 1D). Annexin V/PI staining clearly showed that the apoptotic cell percentage increased from 4.7% in NC transfected cells to about 27% in the miRNA-143 mimics transfected cells (Figure 1E). Taken together, these results indicate that miRNA-143 acts a tumor suppressor in gastric cancer cells.
miRNA-143 suppresses the migration and invasion of the gastric cancer cells
The effects of miRNA-143 were also examined on the migration and invasion of the AGS gastric cancer cells by transwell chamber assay. The results showed that the migration and invasion of the AGS gastric cancer was significantly (P<0.05) reduced in the miR-143 mimics transfected cells (Figure 2A and 2B). The AGS cell migration was inhibited by 46% and the invasion was inhibited by 52% in miRNA-143 transfected cells relative to the NC-transfected AGS cells. Taken together, these results show that miRNA-143 overexpression causes decline in the migration and invasion of the AGS gastric cancer cells.
Figure 2.
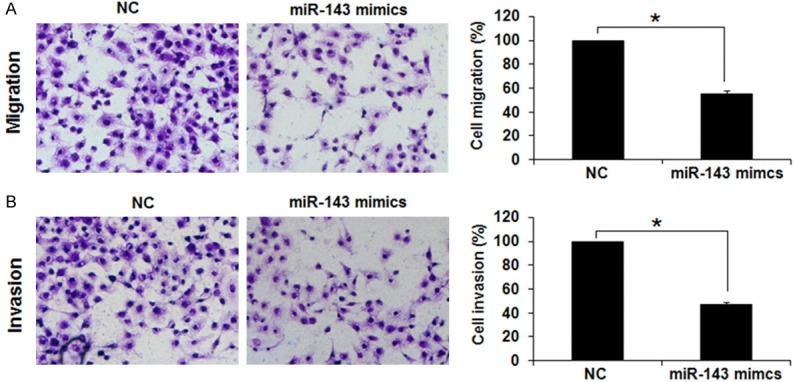
Overexpression of miR-143 inhibits the migration and invasion of gastric cancer cells (A) Transwell assay showing (A) % migration (B) invasion in NC or miR-143 mimics transfected AGS cells. The experiments were performed in triplicate and expressed as mean ± SD (*P<0.05).
miRNA-143 targets STAT3 in gastric cancer cells
The target of miRNA-143 in gastric cancer cells was identified by online TargetScan analysis. STAT-3 was identified as the potential target of miRNA-143 in AGS gastric cancer cells (Figure 3A) and therefore the expression levels of STAT3 were investigated all the gastric cancer cell line as well as the normal cell line. It was found that relative to the expression of miRNA-143 in the normal GES-1 cells, the expression of miRNA-143 was significantly upregulated (upto 3.5 fold) in the gastric cancer cell lines (Figure 3B). However, as the AGS cells were transfected with the miRNA-143 mimics, the expression of STAT3 was considerably downregulated as depicted by the western blot analysis (Figure 3C). Further the dual luciferase assay further confirmed STAT3 to be the target of miRNA-143 in gastric cancer cells (Figure 3D).
Figure 3.
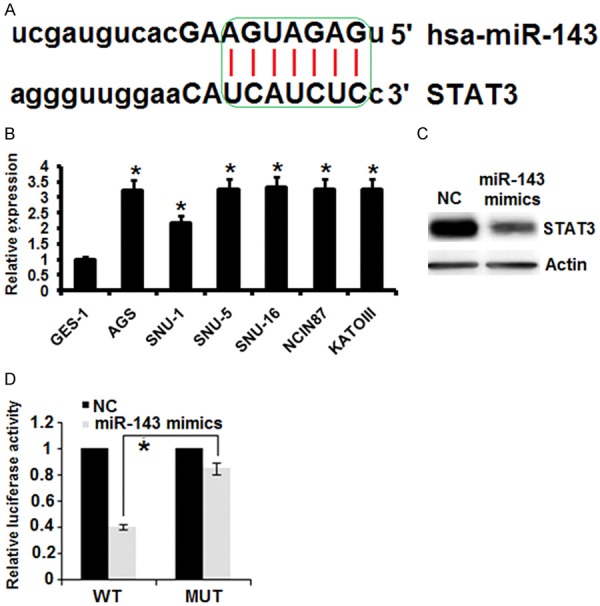
miR-143 exerts its effects by targeting STAT-3. A. TargetScan analysis showing STAT3 as the target of miR-143. B. Expression of STAT in normal GES-1 and six different gastric cancer cell lines. C. Western blot analysis showing the expression of STAT-3 in NC or miR-143 transfected gastric cancer cells. D. Dual luciferase reporter assay. The experiments were performed in triplicate and expressed as mean ± SD (*P<0.05).
Next, we sought to know the effects of the STAT3 silencing on the proliferation, migration and invasion of the gastric cancer AGS cells. Therefore, the AGS cells were transfected with Si-NC or Si-STAT3 and the silencing of expression of STAT3 expression was confirmed by qRT-PCR analysis which showed 4.5 fold downregulation of the STAT3 expression in Si-STAT3 transfected cells relative to the Si-NC transfected cells (Figure 4A). Further, the results of the CCK-8 showed that Si-STAT3 transfection (ie., STAT3 silencing) caused significant (P<0.05) decline in the proliferation, migration and invasion of the AGS gastric cancer cells (Figure 4B-D). These inhibitory effects were almost identical to that of miRNA-143 overexpression.
Figure 4.
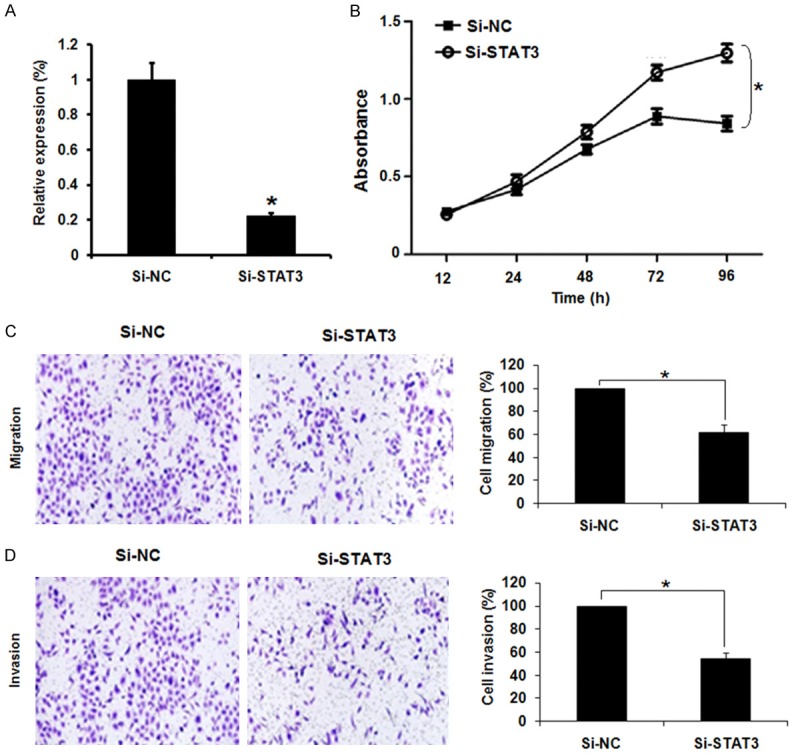
Silencing of STAT-3 inhibits the proliferation, migration and invasion of gastric cancer cells. (A) Expression of STAT-3 in Si-NC or Si-STAT3 transfected AGS cells. (B) CCK-8 assay showing % viability of the Si-NC or Si-STAT3 transfected AGS cells. (C) Transwell assay showing % migration and (D) % invasion of the Si-NC or Si-STAT3 transfected AGS cells. The experiments were performed in triplicate and expressed as mean ± SD (*P<0.05).
Inhibition of STAT3 is essential for tumor suppressive effects of miR-143 in AGS cells
Since, miRNA-143 overexpression and STAT3 silencing exhibited similar effects on the proliferation, migration and invasion of the AGS cells, we sought to know if STAT3 overexpression could nullify the effects of miRNA-143 overexpression in AGS cells. Interestingly, it was found that STAT3 overexpression in the miRNA-143 mimics transfected AGS cells promoted the proliferation, migration and invasion of the AGS cells indicating that the inhibitory effects of the miR-143 overexpression are mainly due to STAT3 suppression (Figure 5A and 5B). On the other hand, inhibition of the miRNA-143 had no effects on the proliferation, migration and invasion of the Si-STAT3 transfected cells further indicating that STAT3 inhibition is essential for the tumor suppressive effects of miRNA-143 (Figure 6A and 6B).
Figure 5.
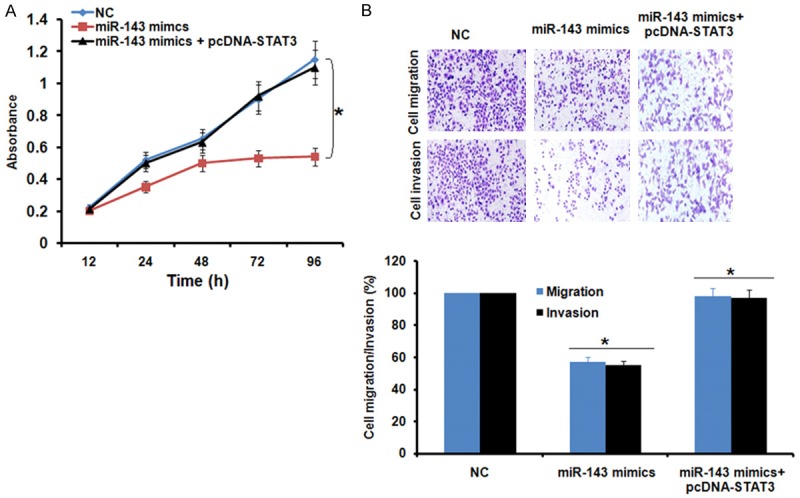
(A) Effect of STAT3 overexpression on the inhibitory effects of miR-143 overexpreson the proliferation and (B) migration and invasion of the AGS cells. The experiwere performed in triplicate and exas mean ± SD (*P<0.05).
Figure 6.
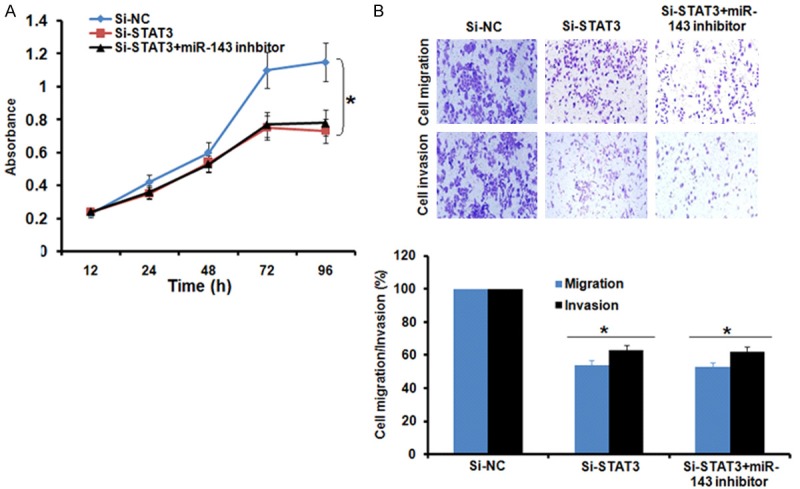
(A) Effect of miR-143 inhibition on the inhibitory effects of STAT3 silencing on proliferation and (B) migration and invasion of the AGS cells. The experiments were performed in triplicate and expressed as mean ± SD (*P<0.05).
Discussion
Although the overall survival rates of gastric cancer have improved, it is still responsible for significant mortality across the globe [16]. For instance, half of the cancer cases detected in East Asian countries are gastric cancers [17]. The currently used drugs for gastric cancer exhibit negative effects on the health of the patients. Additionally, the efficient therapeutic targets for gastric cancer treatment are lacking [17]. Because of the implications of miRNAs in cellular and physiological processes, previous studies have revealed the potential of miRNAs as therapeutic targets for cancer treatment [18,19]. Herein, the role and therapeutic potential of the miRNA-143 was investigated in gastric cancer. It was found that miRNA-143 is aberrantly downregulated in the gastric cancer cells. Previous studies have also shown that miRNA-143 is suppressed in breast cancer cells, confirming the results of the present study [7]. Overexpression of miRNA-143 in the AGS gastric cancer cells caused significant reduction in the proliferation rate of the AGS gastric cancer cells via induction of apoptotic cell death. Furthermore, it also resulted in suppression of migration and invasion of the AGS cells. A number of studies carried out earlier on miRNA-143 have also shown similar results. For example, miRNA-143 could suppress the growth and metastasis of oral cancer cells [20]. In addition, it could also enhance the chemosensitivity of bladder carcinoma cells [21]. Bioinformatic analysis together with dual luciferase indicated STAT3 to be the potential target of miRNA-143. Studies have indicated the involvement of STAT proteins in the initation of proliferative and anti-apoptotic pathway and as such they have been attributed to maintenance of growth and tumorigenisis of several cancer types [22,23]. Herein, we observed that STAT3 is highly upregulated in the gastric cancer and miRNA-143 overexpression could suppress the expression of STAT3. STAT3 silencing could also inhibit the growth of AGS gastric cancer cells similar to that of miRNA-143 overexpression. Furthermore, STAT3 inhibition was found to be essential for the tumor suppressive effects of miRNA-143 on gastric cancer cells. Taken together, miRNA-143 acts a tumor suppressor in gastric cancer and may prove as an essential therapeutic target for gastric cancer.
Quan G, Li J. Circular RNAs: biogenesis, expres- sion and their potential roles in reproduction. J Ovarian Res 2018; 11: 1-12.
Disclosure of conflict of interest
None.
References
- 1.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 4.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, Dougherty U, Robinson V, Mustafi R, Pekow J, Kupfer S, Li YC, Hart J, Goss K, Fichera A, Joseph L, Bissonnette M. EGFR signals downregulate tumor suppressors miR-143 and miR-145 in Western diet-promoted murine colon cancer: role of G 1 regulators. Mol Can Res. 2011;9:960–75. doi: 10.1158/1541-7786.MCR-10-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci. 2015;19:3403–3411. [PubMed] [Google Scholar]
- 7.Xia C, Yang Y, Kong F, Kong Q, Shan C. MiR-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie. 2018;147:98–104. doi: 10.1016/j.biochi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Venerito M, Vasapolli R, Rokkas T, Malfertheiner P. Gastric cancer: epidemiology, prevention, and therapy. Helicobacter. 2018;23(Suppl 1):e12518. doi: 10.1111/hel.12518. [DOI] [PubMed] [Google Scholar]
- 9.Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: epidemiology, prevention, classification, and treatment. Cancer Mang Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, Lanyon G, Martin M, Fraumeni JF Jr, Rabkin CS. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 11.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–13. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fock KM. The epidemiology and prevention of gastric cancer. Aliment Pharmacol Ther. 2014;40:250–260. doi: 10.1111/apt.12814. [DOI] [PubMed] [Google Scholar]
- 13.Smyth EC, Verheij M, Allum W, Cunningham D, Cervantes A, Arnold D ESMO Guidelines Committee. Gastric cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(Suppl 5):v38–v49. doi: 10.1093/annonc/mdw350. [DOI] [PubMed] [Google Scholar]
- 14.Steibel JP, Poletto R, Coussens PM, Rosa GJ. A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics. 2009;94:146–152. doi: 10.1016/j.ygeno.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Hua F, Li CH, Chen XG, Liu XP. Daidzein exerts anticancer activity towards SKOV3 human ovarian cancer cells by inducing apoptosis and cell cycle arrest, and inhibiting the Raf/MEK/ERK cascade. Int J Mol Med. 2018;41:3485–3492. doi: 10.3892/ijmm.2018.3531. [DOI] [PubMed] [Google Scholar]
- 16.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–625. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riboli E. Nutrition and cancer: background and rationale of the European prospective investigation into cancer and nutrition (EPIC) Ann Oncol. 1992;3:783–791. doi: 10.1093/oxfordjournals.annonc.a058097. [DOI] [PubMed] [Google Scholar]
- 18.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 19.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Transl Res. 2011;157:216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Peschiaroli A, Giacobbe A, Formosa A, Markert EK, Bongiorno-Borbone L, Levine AJ, Candi E, D’alessandro A, Zolla L, Finazzi Agrò A, Melino G. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32:797–802. doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 21.Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, Huang L. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181:1372–1380. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 22.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of postimplantationepiblast to pluripotent embryonic stem cells. Nature. 2019;461:1292–5. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bromberg J, Darnell JE Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]


