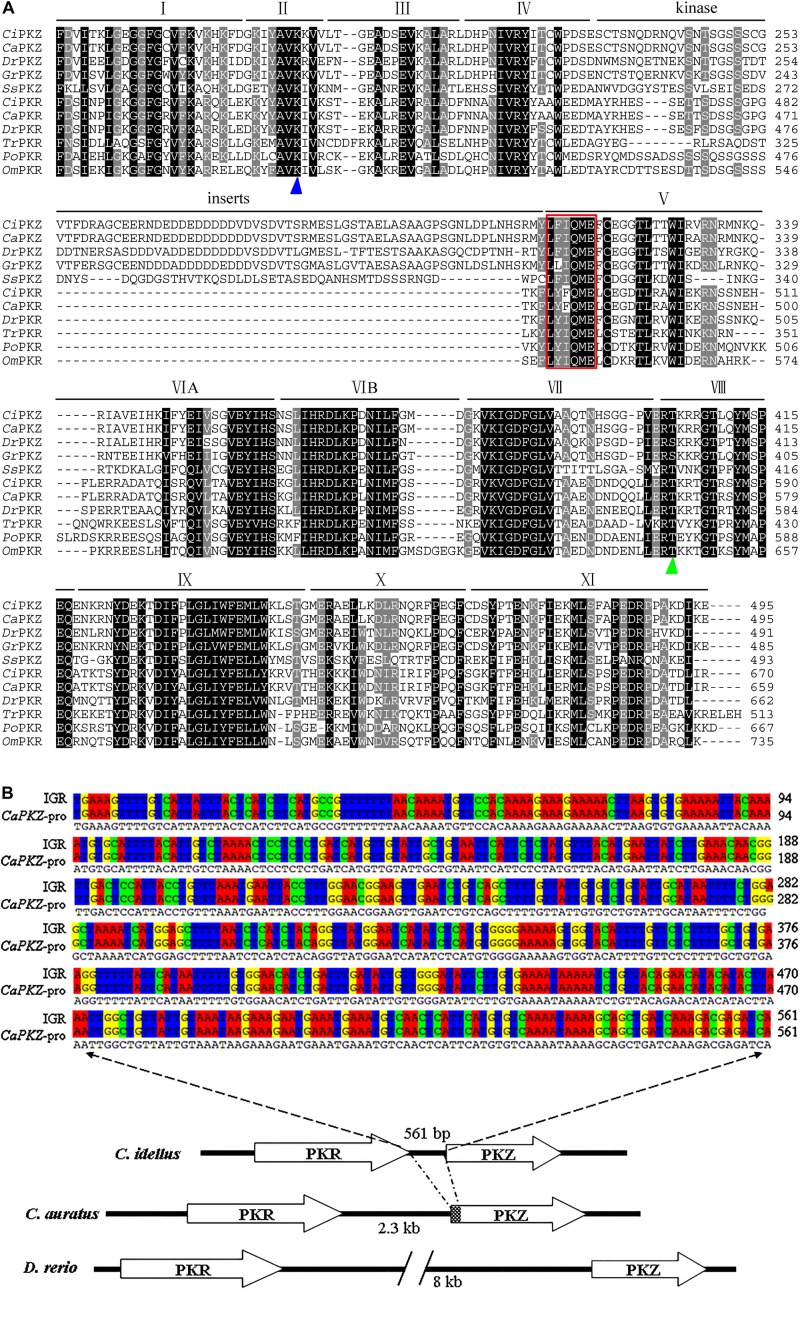
FIGURE 1.
Multiple sequence alignment and genomic arrangement of fish PKR and PKZ genes. (A) Multiple sequence alignment of the kinase domains of fish PKR and PKZ by Clustal X 2.0 program. Residues Lysine (K) for PKR/PKZ catalytic activity (blue triangle) and Serine/Threonine for autophosphorylation (green triangle) are marked under the sequences. The conserved sequence of LFIQME(Y/F)C(D/E) in subdomain V is surrounded by a red box. Identical (shaded in black) and similar (shaded in gray and light gray) residues are indicated. The following abbreviations were used: Ci, Ctenopharyngodon idellus; Ca, Carassius auratus; Dr, Danio rerio; Ss, Salmo salar; Gr, Gobiocypris rarus; Tr, Takifugu rubripes; Po, Paralichthys olivaceus; Om, Oncorhynchus mykiss. (B) Genomic arrangement of tandemly arranged known fish PKR and PKZ genes. The genomic arrangement and relative orientation of PKR and PKZ genes in C. idellus, C. auratus, and D. rerio are shown. Arrows indicate the 5’ to 3’ orientation of the genes. The approximate sizes of intergenic regions are indicated below. Two sequence alignment of the intergenic region (IGR) between CiPKR and CiPKZ and the partial sequence of CaPKZ promoter are displayed above.