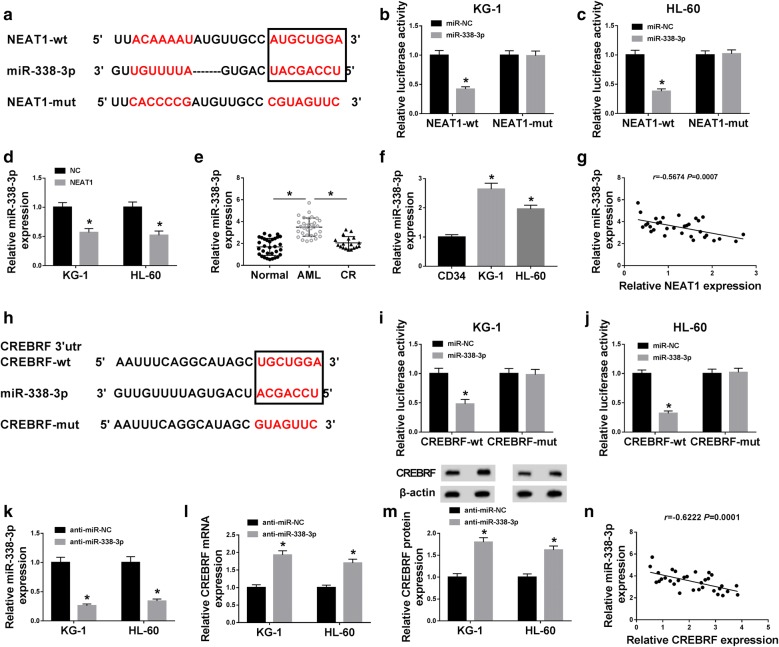
Fig. 5.
NEAT1 negatively interacted with miR-338-3p and miR-338-3p directly targeted CREBRF. a Starbase3.0 was carried out for the prediction of the binding sites between NEAT1 and miR-338-3p. b, c Dual-luciferase reporter assay was used for verifying the combination between NEAT1 and miR-338-3p in AML cells. d The miR-338-3p expression in AML cells transfected with NC or NEAT1 was detected using qRT-PCR. e, f The miR-338-3p expression in AML tissues and cells was assayed by qRT-PCR. g The correlation between NEAT1 and miR-338-3p in AML tissues was analyzed through Spearman’s correlation coefficient. h The bioinformatic analysis between miR-338-3p and CREBRF was executed by Starbase3.0 online database. i, j The affirmation of the binding between miR-338-3p and CREBRF in AML cells was affirmed by the dual-luciferase reporter assay. k The inhibitory efficiency of anti-miR-338-3p on miR-338-3p expression in AML cells was examined by qRT-PCR. l, m QRT-PCR and Western blot were applied for analyzing the effects of anti-miR-338-3p on CREBRF mRNA and protein levels. n The analysis of the relationship between miR-338-3p and CREBRF in AML tissues was administrated by Spearman’s correlation coefficient. *P < 0.05