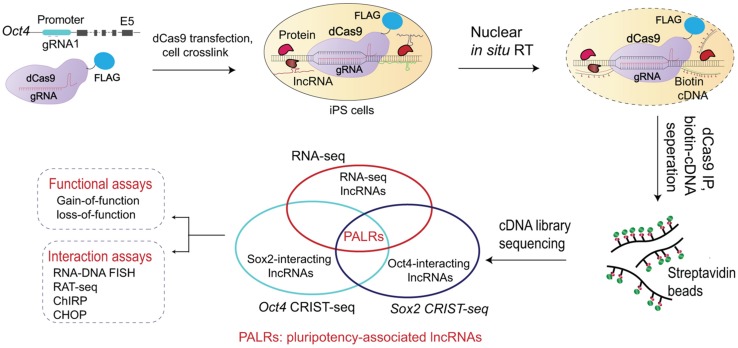
FIGURE 2.
Mapping stemness gene promoter-interacting lncRNA by CRIST-seq. The CRIST-seq assay combines the specificity of Cas9 gene targeting with the simplicity of biotin-lncRNA labeling. Target cells are first transfected with lentiviruses carrying dCas9-FLAG/gRNA that target the promoter of stemness genes, like Oct4 and Sox2, that are two core stem cell factors essential for the establishment and maintenance of pluripotency. The cells are crosslinked to fix the promoter chromatin DNA-lncRNA complex. Following lysis of the cell membrane, the promoter-interacting RNAs are reverse transcribed in situ into biotin-cDNAs (cDNAs) in the isolated nuclei with biotin-dCTP. The promoter chromatin-cDNA complex is immunoprecipitated by a Cas9-FLAG antibody and the promoter-interacting biotin-cDNAs are separated by the biotin-streptavidin bead purification. The CRIST-captured cDNAs are used for library construction and Illumina sequencing is performed for identifying the lncRNAs that interact with the promoter of stemness genes. Integration of the Oct4-Sox2 CRIST-seq data with the RNA-seq data allows the identification of lncRNAs that not only interact with core factor promoters of stem cells, but are also differentially expressed during reprograming. The lncRNA-promoter interaction is subsequently validated by other tools, including RNA-DNA FISH, RAT, ChIRP, and CHOP assays. Gain- and loss-of-function assays are then used to characterize the role of identified lncRNAs in stem cells. The CRIST-seq method offers a more efficient strategy for exploring the lncRNA network in the regulation of reprograming and pluripotency.