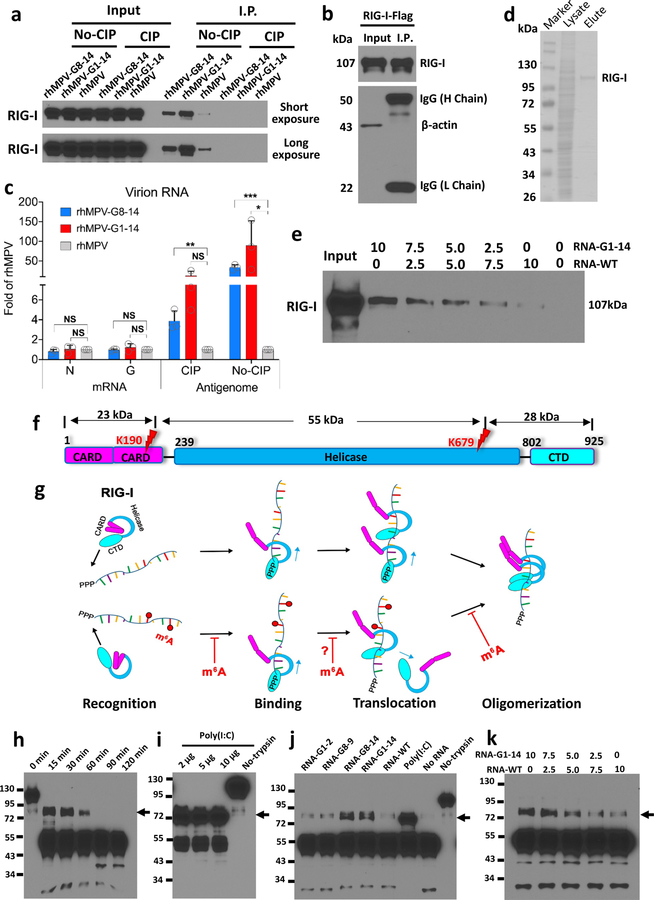
Figure 5. m6A-deficient virion RNA increases RIG-I binding affinity and facilitates RIG-I:RNA conformation change.
(a) Biotinylated virion RNA pulldown RIG-I. Biotinylated virion RNA was conjugated to Streptavidin beads and incubated with A549 cell lysate containing overexpressed RIG-I. The pull-down RIG-I protein was detected by Western blot. (b and c) RIG-I pulldown hMPV RNA. RIG-I conjugated magnetic beads were incubated with virion RNA, N or G mRNA. One aliquot of beads was subjected for Western blot (b). RNA bound to magnetic beads was quantified by real-time RT-PCR (c). (d) Purified Flag-tagged RIG-I protein. (e) Competitive binding of WT virion RNA and m6A-deficient virion RNA to RIG-I. Streptavidin beads-bound rhMPV-G1-14 and rhMPV RNA were mixed at different ratios and incubated with RIG-I protein in the presence of AMP-PNP. RIG-I pulldown was detected by Western blot. (f) Domain structure of RIG-I protein. CARD, caspase activation and recruitment domains; Helicase, helicase domain; CTD, C-terminal domain. Red flashes indicate trypsin cleavage sites. (g) Model for mechanisms of enhanced RIG-I-mediated IFN signaling by m6A-deficient hMPV RNA. RIG-I is in an autorepressed conformation in the absence of ligand. RIG-I CTD recognizes and binds to 5’triphosphate of RNA. 5’triphosphate RNA without m6A has a higher binding affinity to helicase domain of RIG-I. RIG-I is an RNA translocase, moving from 5’-ppp to RNA chain. Internal m6A may serve as a “brake” to prevent RIG-I translocation (indicated by question mark). The RIG-I helicase domain binds the RNA, triggering RIG-I conformational change and subsequent oligomerization. RNAs without m6A more easily induce RIG-I conformational change. The released CARDs of the activated RIG-I:RNA complex are ubiquitinated for downstream signaling. (h-k) Analysis of RIG-I:RNA conformation by limited trypsin digestion. Limited trypsin digestion of RIG-I protein in the absence of RNA ligand for 0–2 h (h), or in the presence of poly (I:C) (i) or virion RNA (j) for 2h was shown. (k) Competition assay. RIG-I incubated with mixtures containing different ratios of RNA of rhMPV-G1-14 and rhMPV, and digested by trypsin for 2h. RIG-I fragments were detected by Western blot. Black arrows in h–k indicate the 80 kDa trypsin-resistant protein band. Western blots (a, b, e, h-k) and SDS-PAGE (d) shown are the representatives of n = 3 biologically independent experiments. Data shown (c) are means of n =3 independent experiments ± standard deviation. Statistical significance was determined by two-sided student’s t-test. *P<0.05; **P<0.01; ***P<0.001.