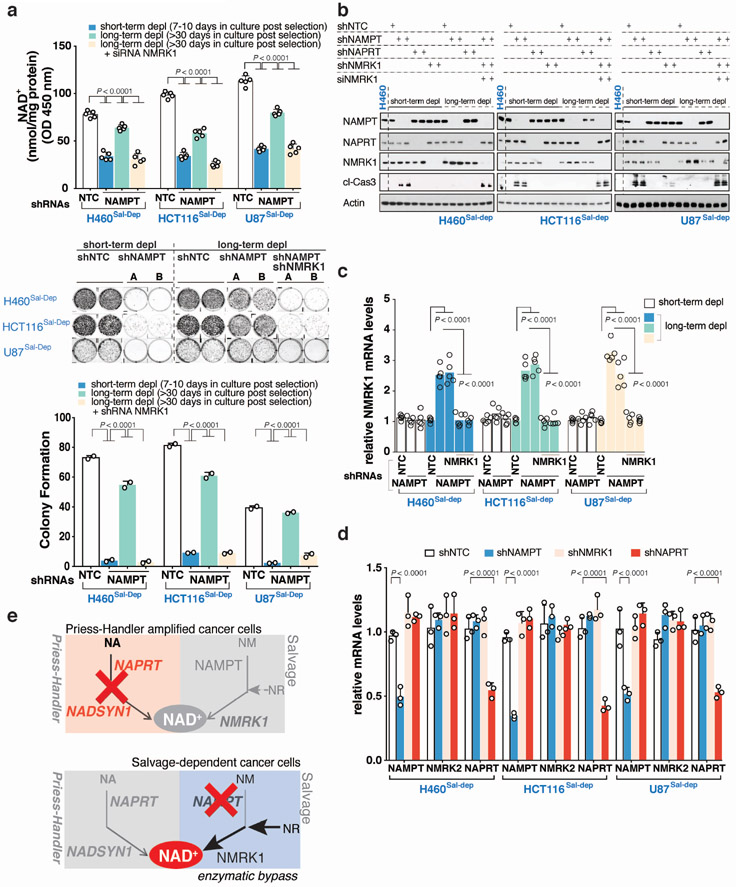
Extended Data Fig. 8: NAMPT deficiency leads to enzymatic bypass of the Salvage-pathway successfully reprogramming NAD biosynthesis in Cancer.
Genetically engineered Salvage-dependent cancer cells including, H460Sal-dep, HCT116Sal-dep and U87Sal-dep were transduced with stable a silencing system targeted against NAMPT using shRNA followed up with puromycin selection. Two different shRNAs were tested against NAMPT. A non-targeting shNTC was used a control. a. Intracellular NAD+ measurement (top). Representative images of clonogenic survival assay using Crystal violet staining (middle) from one of the two independent experiments. Both biological replicates showed similar results. Quantification of colony formation units (bottom). For ‘short-term depl’ cells were seeded 7-10 days post transduction/selection for clonogenic survival assay, while for ‘long-term depl’, cells were seeded ≥30 days post transduction/selection. Cells were stained with crystal violet after 15-18 days of seeding. Salvage-dependent cancer cells stably silenced for NAMPT grown for an extended duration of time (long-term depl), were later silenced for NMRK1 using shRNA. b. Immunoblotting for cleaved-caspase 3 as a measure of cell-death and to test for protein abundance for NAMPT, NMRK1 and NAPRT. Representative blots are from one of the two independent experiments. Both biological replicates showed similar results. Actin was used as a loading control. Salvage-dependent cancer cells stably silenced for NAMPT and grown for an extended duration of time (long-term depl), were later silenced for NMRK1 using siRNA. For ‘short-term depl’ cells were harvested for protein extraction 7-10 days post transduction/selection, while for ‘long-term depl’, cells were harvested for protein extraction ≥30 days post transduction/selection (including transient transfection using siRNA) and later immunoblotted. c. Relative NMRK1, d. Relative NAMPT, NMRK2 or NAPRT transcript levels as measured by quantitative PCR. For ‘short-term depl’, cells were harvested for RNA extraction 7-10 days post transduction/selection, while for ‘long-term depl’ cells were harvested for RNA ≥30 days post transduction/selection. Salvage-dependent cancer cells stably silenced for NAMPT and grown for an extended duration of time (long-term depl), were later silenced for NMRK1 using shRNA. e. Schematic overview of the model illustrating NAD pathway addiction in cancer is driven through two separate mechanisms, one that gets shaped by gene amplification (left) while the other through epigenetic reprogramming (right). The model demonstrates tissue context-based amplifications of genes encoding key enzymes (NAPRT/NADSYN1) of the PH-pathway and subsequent tumor cell dependence that is absolute and not subjected to enzymatic bypass rewiring. In contrast, epigenetically-determined dependence on the NAMPT driven Salvage-pathway is subject to enzymatic bypass, requiring combination therapies. Data are representative of five (a-top,c, n=5), three (d, n=3) and two (a-middle, a-bottom, n=2) independent biological replicates. Data as scatter plots with bars are represented as mean ± s.d, analysed by one-way ANOVA with Tukey’s multiple comparisons test (a,c,d). For gel source data, see Supplementary Fig. 1.