Fig. 1: The eukaryotic ribosome-associated quality control pathway.
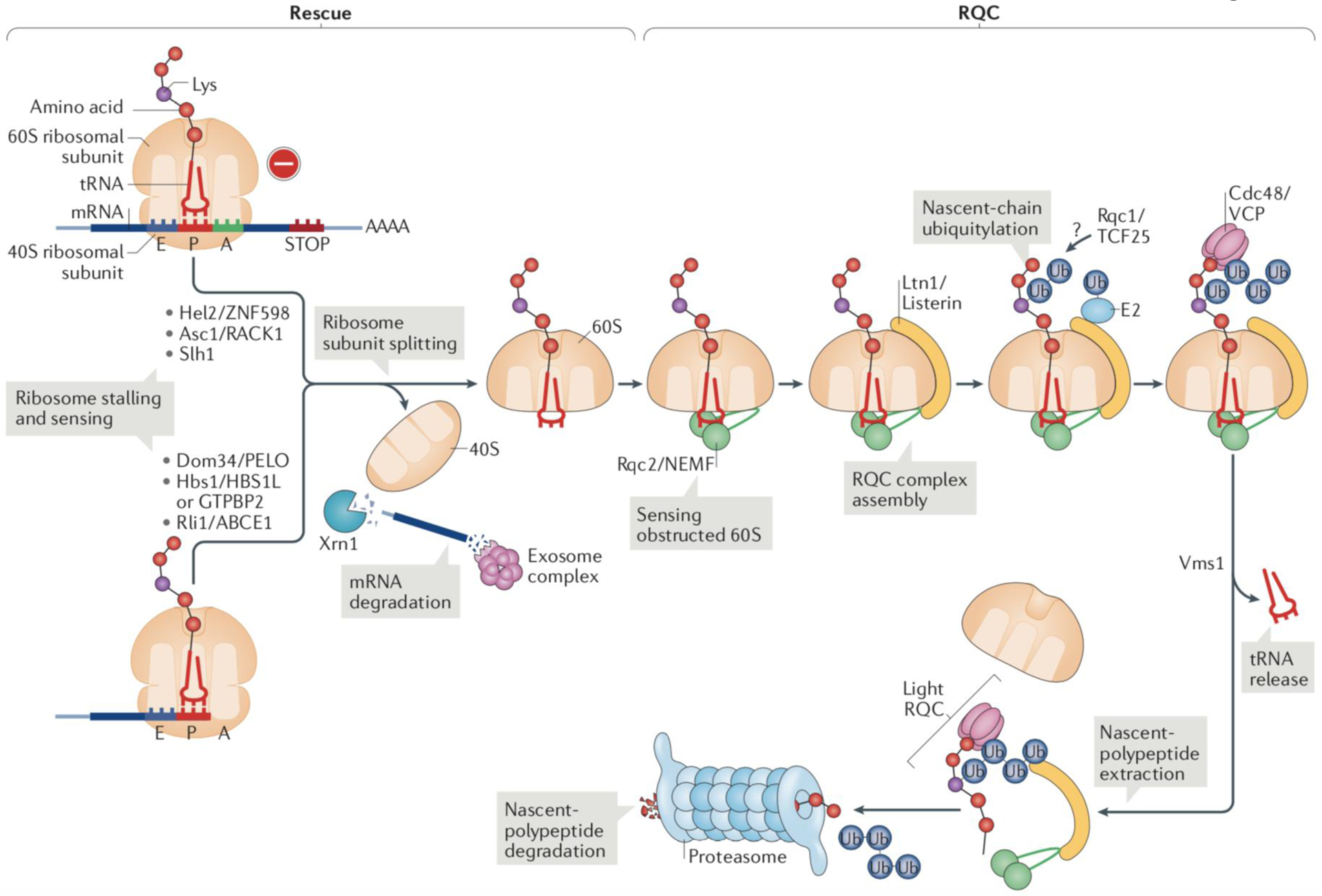
The ribosomal rescue and Ribosome-Associated Quality Control (RQC) pathways for protein surveillance consist of two sequential steps that each sense a unique defect. The first step (rescue) senses stalled ribosomes and mediates 80S subunit splitting; the second step (RQC) detects 60S subunits obstructed with peptidyl-tRNA—products of the rescue step—and promotes the resolution of this aberrant structure, to release free, translation-competent 60S ribosomal subunits and mediate proteolysis of the nascent chain.
(Left) Ribosomal rescue. Ribosomes that are stalled at the mRNA 3’ end are sensed by Hbs1 in yeast (or HBS1L and GTPBP2 in mammals) and Dom34 (pelota (PELO) in mammals). The mechanisms for the recognition of ribosomes stalled on internal mRNA sequences are poorly understood and at least in some cases involve the E3 ubiquitin-protein ligase Hel2/ZNF598. Whether Dom34/PELO also acts downstream of Hel2/ZNF598 is not known, but these two factors can target the same mRNA molecule, as ribosome stalling elicits endonuclease cleavage of the transcript4. Dom34/PELO recruits Rli1 (ATP-binding cassette protein ABCE1 in mammals), which separates the small (40S) and large (60S) ribosomal subunits (Box 1). The released truncated mRNA is degraded by the 5’−3’ exoribonuclease Xrn1 and the exosome complex, which prevents the mRNA from being translated again, as it may be defective. However, unlike eRF1, Dom34 lacks peptidyl-tRNA hydrolase activity, the peptidyl-tRNA remains bound to the 60S subunit. Although the peptidyl-tRNA−60S complex can potentially reassociate with free 40S subunits, the reassociation can be reversed by Dom34, Hbs1 and Rli113,14,115.
(Right) RQC. Resolution of the peptidyl-tRNA−60S complex is initiated by binding of the Rqc2 (NEMF in mammals) subunit of the RQC complex, which recruits and stabilizes the binding of the E3 ligase Ltn1/Listerin. At this stage, Rqc2 may synthesize CAT tails to help expose Lys residues that are buried in the ribosomal exit tunnel, to be ubiquitylated by Ltn1/Listerin (see below). The ribosome binding mode of the Rqc1 subunit (TCF25 in mammals) of the RQC complex and its exact function remain unknown. The ubiquitin chain that is polymerised by Ltn1 on the nascent polypeptides signals recruitment of the AAA ATPase Cdc48 and its cofactors. Cdc48 extracts nascent polypeptides from the 60S ribosomal subunit after they have been released from the conjugated tRNA by Vms1 (ANKZF1 in mammals). Released polypeptides can then be degraded by the proteasome. The extracted nascent polypeptides may remain associated with RQC subunits in the form of a ‘light RQC’ complex before proteasomal degradation.
