Fig. 3.
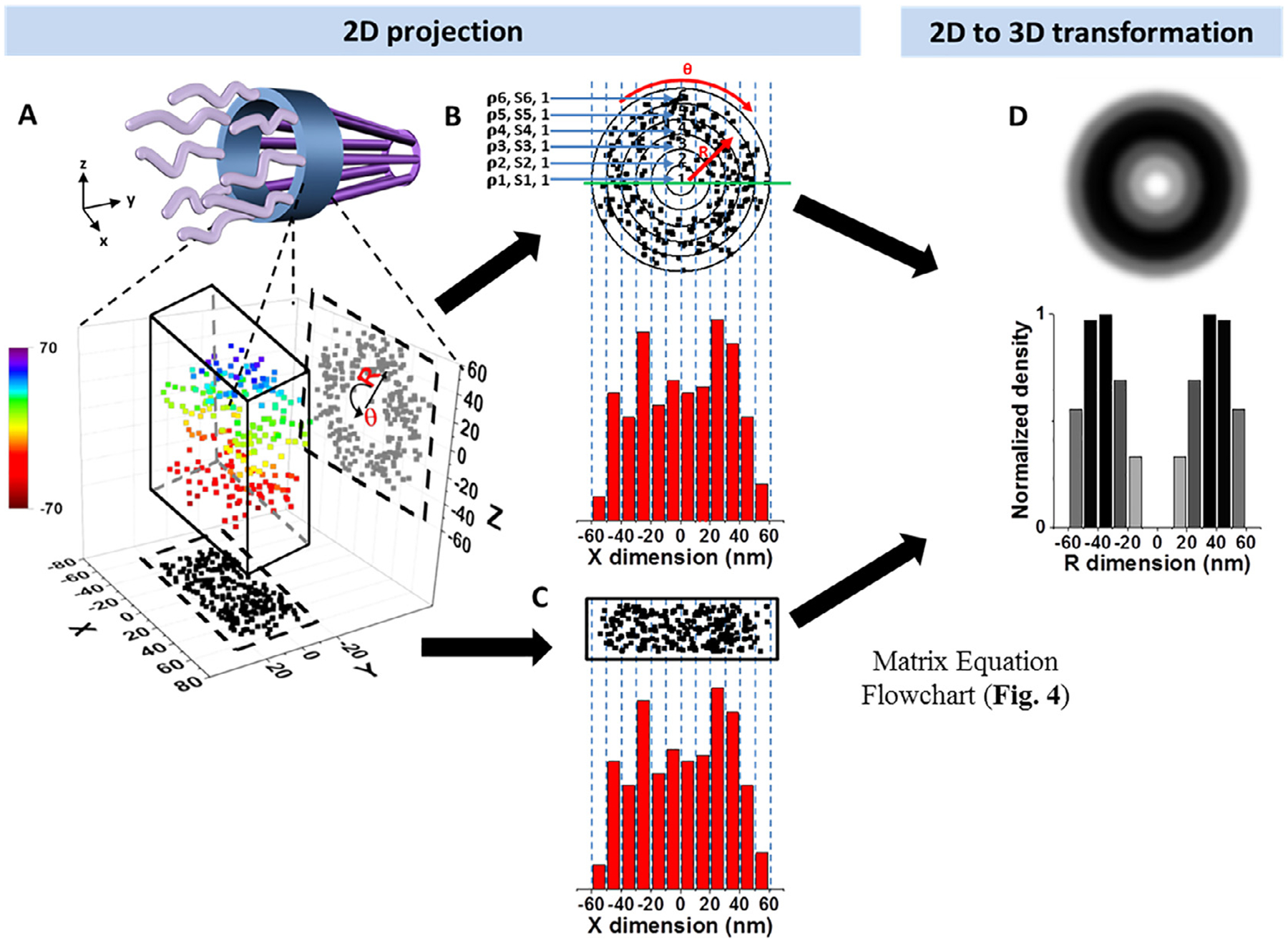
A schematic demonstration, with simulated data, of the 2D to 3D transformation algorithms for molecules that diffuse through the NPC. (A) 3D spatial locations of randomly diffusing molecules inside the NPC can be coordinated in a cylindrical coordination system (R, Ø, Y) due to the cylindrical rotational symmetry of the NPC. The 3D molecular locations in the NPC (rainbow colored for Z position) are projected onto a 2D plane in a Cartesian coordination system (X and Y, shown as black points) by microscopy imaging (X and Z shown as gray points). (B) The cross-sectional view of all the locations shown in Fig. 3A (same as the gray points from the X and Z dimension). These locations can be grouped into the sub-regions between concentric rings. Given the high number of randomly distributed molecules in the NPC the spatial density of locations (ρi) in each sub-region (s(i,j)) between two neighboring rings will be rotationally symmetrical and uniform. These locations can be further projected into 1D along the X dimension. The locations along X dimension can be clustered in a histogram with j columns. The total number of locations in each column (A(i,j)) is equal to , which is further explained in Fig. 4. (C) Histograms of 2D projected data from microscopy experiments are identical to B, thereby allowing us to use the aforementioned formula to determine the density of each concentric ring. (D) Using the algorithms, 2D projected data can be used to reconstruct the 3D spatial distribution of proteins traveling through the NPC.
