SUMMARY
Botulinum neurotoxin (BoNT) is one of the most acutely lethal toxins known to humans, and effective treatment for BoNT intoxication is urgently needed. Single-domain antibodies (VHH) have been examined as a countermeasure for BoNT because of their high stability and ease of production. Here, we investigate the structures and the neutralization mechanisms for six unique VHHs targeting BoNT/A1 or BoNT/B1. These studies reveal diverse neutralizing mechanisms by which VHHs prevent host receptor binding or block transmembrane delivery of the BoNT protease domain. Guided by this knowledge, we design heterodimeric VHHs by connecting two neutralizing VHHs via a flexible spacer so they can bind simultaneously to the toxin. These bifunctional VHHs display much greater potency in a mouse co-intoxication model than similar heterodimers unable to bind simultaneously. Taken together, our studies offer insight into antibody neutralization of BoNTs and advance our ability to design multivalent anti-pathogen VHHs with improved therapeutic properties.
Graphical Abstract
In Brief
Botulinum neurotoxins (BoNTs) are extremely toxic biothreats. Lam et al. report the crystal structures and neutralizing mechanisms of six unique antitoxin VHHs against BoNT/A1 and BoNT/B1, the two major human pathogenic BoNTs. They then develop a platform for structure-based rational design of bifunctional VHH heterodimers with superior antitoxin potencies.
INTRODUCTION
Botulinum neurotoxins (BoNTs) are the most potent toxins to humans. BoNT exposure inhibits the release of acetylcholine in presynaptic neurons, leading to a flaccid neuromuscular paralysis that causes death by respiratory collapse. There are seven classical BoNT serotypes (designated A through G), with several new BoNT or BoNT-like serotypes identified within the past several years (Tehran and Pirazzini, 2018). BoNT/A, /B, /E, and /F are the etiological sources of most cases of endemic human botulism. Although naturally occurring botulism is rare, BoNTs can be misused as a bioweapon and, thus, have been classified as tier 1 select agents by the Centers of Disease Control and Prevention (CDC). BoNT/A and BoNT/B are also increasingly used therapeutically for the treatment of numerous medical conditions, thereby creating the accompanying risk of iatrogenic botulism.
Structurally, each BoNT molecule is composed of a light chain (LC; the protease domain) and a heavy chain (HC) comprised of an N-terminal translocation domain (HN) and a C-terminal receptor-binding domain (HC). Functionally, HC determines neuronal specificity by recognizing a polysialoganglioside (e.g., GT1b) and a protein receptor, synaptotagmin (Syt) I/II (for BoNT/B, /G, and /DC) or glycosylated synaptic vesicle protein 2 (SV2) (for BoNT/A, /D, /E, and /F), located on the presynaptic membrane (Chai et al., 2006; Jin et al., 2006; Montecucco, 1986; Stenmark et al., 2008; Yao et al., 2016). HC of BoNT/B, /G, and /DC additionally carries a hydrophobic loop, termed the HC-loop, which interacts with host membrane lipids (Stern et al., 2018; Zhang et al., 2017; Figure 1A). Under acidic conditions, the HN undergoes a pH-induced structural rearrangement and forms a protein channel that delivers the unfolded LC to the cytosol (Fischer et al., 2012; Koriazova and Montal, 2003; Lam et al., 2018; Montal, 2009). The translocated LC then cleaves cytosolic SNARE proteins, thereby blocking neurotransmitter release and nerve transmission (Agarwal et al., 2009; Breidenbach and Brunger, 2004).
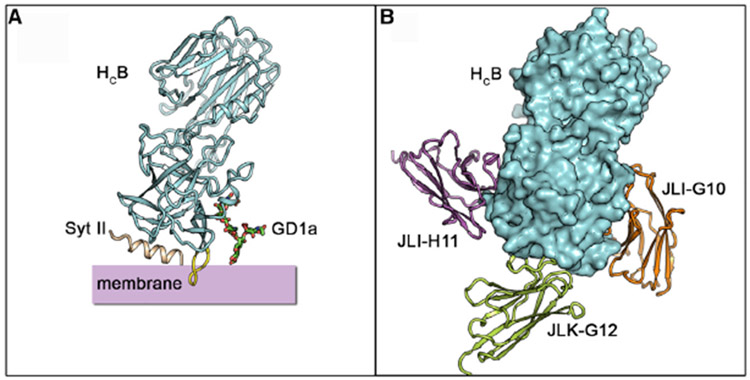
Figure 1. Structures of HCB in Complex with JLI-G10, JLK-G12, or JLI-H11.
(A) A model illustrating the binding of HCB to ternary receptors: Syt II, disialoganglioside 1a (GD1a), and lipid membrane.
(B) A model of HCB simultaneously bound with three VHHs. HCB is positioned in the same orientation as in (A).
Currently, the only available antitoxin remedies are polyclonal antibodies from horse or human serum, which have known health risks and are in limited supply (Schussler et al., 2017). Monoclonal antibodies (mAbs) against BoNT/A have been developed under phase I/II clinical trials (Espinoza et al., 2019; Nayak et al., 2014). Small proteins such as heavy-chain-only camelid antibodies (called VHHs, nanobodies, or single-domain antibodies) and de novo designed mini-proteins against the toxins are currently being developed as alternatives (Chevalier et al., 2017; Conway et al., 2010; Godakova et al., 2019; Mukherjee et al., 2012; Thanongsaksrikul et al., 2010). These small proteins have high stability, can be economically produced, display high binding affinity, and have been shown to function effectively as antitoxins in animal models (Dong et al., 2010; Herrera et al., 2015; Schmidt et al., 2016; Sheoran et al., 2015; Vance et al., 2013; Vrentas et al., 2016). However, the therapeutic applications of these antitoxins have been limited by a lack of understanding of the molecular mechanisms underlying BoNT neutralization, the extreme potency of BoNTs (in vivo lethal blood concentrations at sub-pM), as well as the diverse sequences among different BoNT serotypes and subtypes.
In earlier studies, we found that VHH-based neutralizing agents (VNAs) consisting of VHH heterodimers joined by a flexible peptide linker possessed significantly improved antitoxin potencies (Mukherjee et al., 2012). Similar results have been obtained when developing VNAs for other toxins, such as ricin, anthrax, Shiga toxins, and Clostridium difficile toxins (Herrera et al., 2015; Schmidt et al., 2016; Sheoran et al., 2015; Vance et al., 2013; Vrentas et al., 2016; Yang et al., 2014). We hypothesized that novel heterodimeric VNAs, which are composed of two VHHs connected by a spacer that permits simultaneous binding, could be rationally designed based on structural and mechanistic knowledge of individual VHHs. Such designer VNAs could display greater efficacy due to their substantially improved binding kinetics and possibly complementary neutralizing mechanisms.
Here, we carried out comprehensive structure-function characterization on six neutralizing VHHs that bind non-competitively, three against BoNT/A1 and three against BoNT/B1. Our studies reveal a spectrum of mechanisms by which VHHs attack and neutralize BoNT at different stages of intoxication, such as disrupting receptor binding, inhibiting channel formation, and preventing LC delivery. We then demonstrate that designer VNAs composed of two VHHs with proximal binding epitopes and distinct neutralizing mechanisms have superior potency in a mouse co-intoxication assay over similar VHHs that are unable to bind simultaneously. Together, this study establishes a platform for structure-based rational design of improved antitoxins against BoNTs.
RESULTS
Isolation of BoNT/B1-Neutralizing VHHs
Our initial goal was to develop bivalent VHHs that neutralize the action of both BoNT/A and BoNT/B, which account for 90% of the human botulism cases (Centers for Disease Control and Prevention, 2019). We previously reported the characterization of two panels of VHHs selected for binding to BoNT/A1 and BoNT/B1 (Mukherjee et al., 2012), including several VHHs capable of neutralizing their toxin target. Although we obtained neutralizing VHHs recognizing non-overlapping epitopes on all three domains of BoNT/A1, we obtained only one neutralizing VHH for BoNT/B1. To seek additional neutralizing BoNT/B-binding VHHs, we obtained a new panel of VHHs selected for binding to catalytically inactive BoNT/B1 (ciBoNT/B1). The phage-displayed VHHs were derived from several immunized alpacas and were selected for binding to plastic-coated ciBoNT/B1 or LC/B or binding to ciBoNT/B1 captured by a non-neutralizing VHH (JEQ-H11). The aa sequences of 13 novel BoNT/B1-binding VHHs are shown in Figure S1A. We systematically examined their EC50 binding affinity to ciBoNT/B1, mapped the binding epitopes of these VHHs using ELISA against LC/B and HCB, and tested their ability to neutralize BoNT/B1 intoxication using primary neurons. All these results are summarized in Table S1 and Figure S1B.
Four Neutralizing VHHs Block BoNT/B1 Binding to Host Receptors
Notably, four of the five most potent neutralizing VHHs (JLK-G12, JLO-G11, JLI-G10, and JLI-H11) recognize HCB. In our earlier studies, we identified a potent neutralizing VHH (ciA-C2) against BoNT/A1, which binds to the HC domain of BoNT/A1 and prevents its binding to its protein receptor SV2 (Yao et al., 2017). But, no neutralizing epitope has been mapped to HCB before. To further understand the molecular basis of toxin neutralization by these four HCB-binding VHHs, we determined the crystal structures of HCB in complex with JLK-G12, JLO-G11, JLI-G10, or JLI-H11 at 2.2 to 3.2 Å resolution (Table S2). Structural analysis demonstrated that all four VHHs bind to the C-terminal subdomain (HCC) of HCB, which harbors the binding sites for BoNT/B’s protein receptor Syt and polysialoganglioside (Figure 1B).
VHH JLI-G10 binding to HCB buries a molecular surface area of ~974 Å2 per molecule (calculated by PDBePISA v1.51) (Figure 2A). The structure of the HCB–GD1a (PDB: 4KBB) complex revealed that the polysaccharide head group of a ganglioside makes contacts with a shallow groove in HCB composed of a “E1190 … H1241 … 1260SXWY1263 … G1277” motif, which is also conserved in other BoNT serotypes (Berntsson et al., 2013; Lam et al., 2015; Rummel, 2013). We found that the complementarity-determining region 3 (CDR3) of JLI-G10 occupies the same area on HCB as GD1a, which should strongly compete for ganglioside binding. Specifically, residue R106 of JLI-G10 interacts with E1190, H1241, and W1262 of HCB, and the backbone carbonyl of A104 of JLI-G10 is hydrogen bonded with Y1263 and G1277 of HCB. The VHH-HCB complex is further strengthened by electrostatic and hydrophobic interactions (Figures S2A and S2D). Consistent with the structural finding, JLI-G10 significantly reduced binding of HCB to GT1b-containing liposomes in a co-sedimentation assay (Figures 2B and S7D).
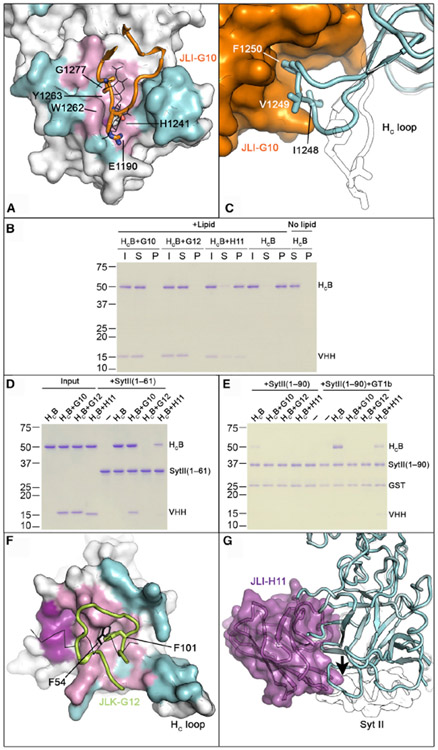
Figure 2. VHHs JLI-G10, JLK-G12, and JLI-H11 Inhibit HCB Binding to Host Receptors.
(A) JLI-G10 occupies the ganglioside-binding pocket on HCB. HCB–JLI-G10 is superimposed with HCB–GD1a (PDB: 4KBB). GD1a is drawn in the line model. The JLI-G10-binding interface is colored cyan, whereas regions overlapping or non-overlapping with the GD1a-binding site are colored pink and magenta, respectively. Key residues on the CDR3 loop (orange illustration) that interact with the conserved ganglioside recognition motif are shown in sticks.
(B) JLI-G10 and JLK-G12 inhibit the binding of HCB to GT1b-containing liposomes. HCB pre-bound with VHHs was incubated with liposomes containing 70/20/10 mol% BrPC/DOPS/GT1b. After liposomes were pelleted, HCB and VHHs in the input (I), supernatant (S), and pellet (P) fractions were analyzed by SDS-PAGE.
(C) JLI-G10 (orange) occupies the HC-loop (cyan illustration) of BoNT/B1. Hydrophobic residues reported to be crucial for lipid interaction are indicated. The structure of the apo BoNT/B1 (PDB: 2NP0) is shown (white illustration) for comparison.
(D) JLK-G12 and JLI-H11, but not JLI-G10, prevent HCB from binding to Syt II (1–61). Pull-down assay was performed using HCB as a prey and the GST-tagged Syt II (1–61) as a bait. After binding, the GST resins were washed three times and the bound proteins were released and subjected to SDS-PAGE.
(E) JLI-G10 and JLK-G12 block the interaction of HCB to its ternary receptors. GST-tagged Syt II (1–90) was used as baits. Pull-down assay was done in a buffer containing 0.5% Triton X-100, with the addition of GT1b as indicated. These experiments were performed in triplicate. Uncropped images of gels and quantification of protein band intensities are shown in Figures S7A-S7F.
(F) JLK-G12 binds at the Syt-binding site on HCB. The JLK-G12- and the Syt II-binding surfaces of HCB are colored cyan and magenta, respectively. The overlapping interface is colored pink. Note that residue F101 in the CDR3 loop of JLK-G12 (illustration) and F54 of Syt II (line) occupy the same hydrophobic pocket on HCB.
(G) Superposition of HCB–JLI-H11 (cyan and purple) and HCB–Syt II complexes (white; PDB: 2NM1). Note that JLI-H11 creates a side-to-side clash (arrow) with the N terminus of Syt II (white illustration). See also Figures S2, S3, and S6.
Interestingly, JLI-G10 causes a large conformational change in the membrane-binding HC-loop (E1245–E1252) on HCB by directly binding to the hydrophobic residues I1248 and F1250 (Figures 2C and S2D). Prior studies showed that deleting I1248-F1250 in this region potently inhibited the binding of HCB to membrane-embedded Syt II, even though these residues are not directly involved in binding the luminal domain of Syt II (Dong et al., 2003; Stern et al., 2018). It, thus, prompted us to hypothesize that JLI-G10 might also impair the association of HCB with the plasma membrane in addition to competing for ganglioside binding. To test this hypothesis, we assessed the ability of the HCB–JLI-G10 complex to interact with the soluble GST-Syt II luminal domain (residues 1–61) or a membrane-anchored GST-Syt II (residues 1–90) that carries the transmembrane domain and is solvated in Triton X-100 micelles. The latter was shown to faithfully mimic Syt II insertion in a membranous environment (Stern et al., 2018). As expected, JLI-G10 did not disturb HCB binding to the soluble GST-Syt II (Figures 2D and S7E). However, JLI-G10 completely blocked the binding of HCB to GST-Syt II (1–90) embedded in Triton X-100 micelles, even in the presence of co-receptor GT1b (Figures 2E and S7F) that should form a cis-complex with Syt II in the detergent micelle to enhance HCB binding (Flores et al., 2019). These data collectively suggest that JLI-G10 inhibits the binding of BoNT/B1 to neuronal membrane by simultaneously occupying the membrane-binding HC-loop and the ganglioside-binding pocket on HCB.
The binding sites of JLK-G12 and JLO-G11 on HCB partially overlap with the Syt II-binding pocket (Jin et al., 2006; Figures 2F, S2, and S3). Because these two VHHs share a high sequence identity of ~84% and bind to HCB in a similar manner (root-mean-square deviation [RMSD] = 0.41 Å), we focused on JLK-G12 for further study, which has a slightly bigger interface with HCB than JLO-G11 (943 Å2 versus 872 Å2). Interestingly, JLK-G12 shares similarities with Syt II when recognizing HCB. For example, a phenylalanine (F101) in the CDR3 loop of JLK-G12 is inserted into the same hydrophobic pocket on HCB that otherwise accommodates F54 of Syt II, which is crucial for high affinity binding of BoNT/B to Syt II (Figure 2F; Jin et al., 2006). This mimicry suggests that JLK-G12 neutralizes BoNT/B1 by blocking its binding to Syt. This is supported by the observation that JLK-G12 clearly prevented the binding of HCB to GST-Syt II (1–61) (Figures 2D and S7E).
Notably, JLK-G12 partly interacts with the HC-loop of HCB, which suggests that JLK-G12 may also interfere with lipid binding of HCB (Figures S2B and S2E). Consistent with this structural finding, we observed that JLK-G12 inhibited binding of HCB to GT1b-containing liposomes in a liposome co-sedimentation experiment, even though it does not directly compete with GT1b binding (Figures 2B and S7D). Furthermore, JLK-G12 also inhibited binding of HCB to detergent-solvated Syt II (1–90) supplemented with GT1b (Figures 2E and S7F). Therefore, JLK-G12 appears to inhibit BoNT/B1 binding to both its neuronal protein receptor Syt and plasma membrane, which leads to potent toxin neutralization.
The neutralizing mechanism of VHH JLI-H11 was initially confusing, because it binds HCB in a region that has no known function (Figures 2G, S2C, and S2F). A detailed structural analysis revealed that the N terminus of JLI-H11 clashes with the N terminus of a fragment of Syt II (residues 44–60) based on the crystal structure of a HCB–Syt II complex. It is worth noting that only residues 44 to 60 of Syt II were resolved in this crystal structure, whereas Syt II residues N-terminal to this fragment were not observed due to high structural flexibility (Jin et al., 2006). Therefore, we hypothesized that JLI-H11 may clash more severely with Syt II in a region N-terminal to fragment 44–60. Supporting this notion, we found that JLI-H11 partially inhibited HCB binding to both soluble Syt II (1–61) and the detergent-solvated Syt II (1–90) (Figures 2D, 2E, S7E, and S7F). As the JLI-H11 epitope does not overlap with the HC-loop or the glycolipid-binding pocket of HCB, the HCB–JLI-H11 complex was able to bind the GT1b-containing liposome and partially interacted with detergent-solvated Syt II supplemented with GT1b (Figures 2B, 2E, S7D, and S7F). Together, these findings suggest that JLI-H11 neutralizes BoNT/B1 by competing with its binding to the protein receptor.
VHH ciA-B5 Blocks Membrane Insertion of BoNT/A1 Translocation Domain
In our earlier work, we obtained a large panel of unique BoNT/A1-binding VHHs recognizing seven non-overlapping epitopes on the toxin (Mukherjee et al., 2012). Among these VHHs, ciA-B5, ciA-C2, and ciA-H7 potently neutralized BoNT/A1 intoxication through binding at unique epitopes. VHH ciA-C2, which binds to the HC domain of BoNT/A1, was recently shown to neutralize BoNT/A1 through inhibition of its binding to the protein receptor SV2 (Yao et al., 2017). Here, we performed structural analyses on ciA-B5 and ciA-H7, which bind to HN and LC of BoNT/A1, respectively. We also included VHH ciA-D12 as a control for ciA-H7 because ciA-D12 binds to a distinct epitope in LC/A and possesses marginal neutralizing activity in cell-based assays (Mukherjee et al., 2012). After extensive crystallization screening trials, we successfully crystallized a hetero-tetrameric complex composed of LCHN/A (residues M1 to K871), including both the LC and the HN of BoNT/A1, and all three VHHs in a 1:1:1:1 ratio. The structure of LCHN/A–B5–D12–H7 was determined at 2.16-Å resolutions (Table S2).
We first focused on the neutralizing mechanism of ciA-B5 that recognizes an epitope on the N-terminal boundary of the rod-like HN (Figure 3A). We found that ciA-B5 inhibited the HNA-mediated dye release of calcein-loaded asolectin liposomes by over 85% (Figure 3B) and potently reduced the ability of HNA to dissipate valinomycin-induced membrane potential in anionic liposomes in a concentration-dependent manner (Figure 3C). These data demonstrated that ciA-B5 blocks the channel formation of HNA, which is a crucial step during BoNT/A intoxication. Interestingly, aa sequence analysis shows that the ciA-B5 epitope on BoNT/A1 (residues 600–616) partially overlaps with a putative channel-forming amphipathic region (Lam et al., 2018; Mansfield et al., 2019) (residues 593–686; Figures 3A, S2G, S2J, and S4A). Specifically, M39 and Y103 of ciA-B5 interact with L604, V607, V611, and Y612 of BoNT/A1, and R60 of ciA-B5 forms a salt bridge with D616 of BoNT/A1. Therefore, ciA-B5 may interfere with membrane insertion of its epitope into the membrane during channel formation.
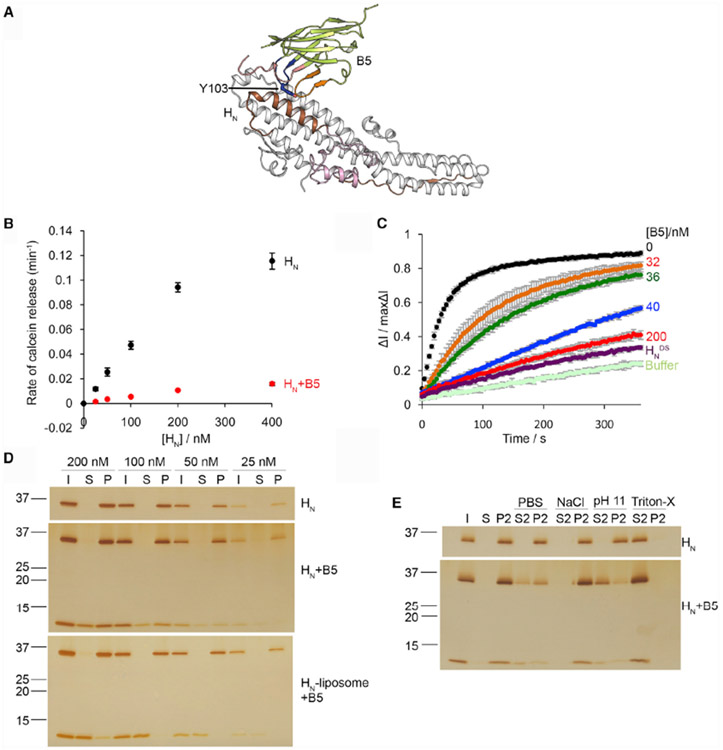
Figure 3. VHH ciA-B5 Inhibits the Channel Formation of HNA.
(A) ciA-B5 occupies the membrane-interacting region on HNA. The putative transmembrane region is colored brown, and the BoNT-switch is highlighted in pink. The CDR1, 2, and 3 of ciA-B5 are colored in red, orange, and blue, respectively.
(B) Calcein dye release assay. HNA(0–400 nM) was tested with liposomes loaded with 50 mM calcein at pH 4.6, in the presence or absence of ciA-B5. The rate of calcein dye release was determined based on the increase of fluorescence at 525 nm during excitation at 493 nm.
(C) Membrane depolarization assay. Liposomes were polarized at a positive internal voltage by adding valinomycin in the presence of a transmembrane KCl gradient. Membrane potential was measured using the voltage-sensitive fluorescence dye ANS. After 3 min, HNA with various concentrations of ciA-B5 was added. The data are presented as ± SD; n = 3.
(D) Liposome co-sedimentation assay. HNA(25–200 nM) or HNA pre-bound with ciA-B5 in 1:1.5 molar ratio was incubated with liposomes containing 80/20 mol% BrPC/DOPS. Alternatively, HNA was incubated with liposomes followed by co-incubation with ciA-B5 before centrifugation. After liposomes were pelleted, HNA and ciA-B5 in the input (I), supernatant (S), and pellet (P) fractions were analyzed by SDS-PAGE.
(E) The pellet fraction of liposomes incubated with 100 nM of HNA or HNA–ciA-B5 was re-suspended in buffers containing either PBS, 1 M NaCl and 50 mM sodium acetate (pH4.6), 0.15 M NaCl and 50 mM CAPS (pH 11), or 2% Triton X-100. The re-suspended liposomes were re-centrifuged and the supernatant (S2) and pellet (P2) fractions were separated. These experiments were performed in triplicate and quantification of protein band intensities are shown in Figures S7L-S7N. Uncropped images of gels are shown in Figures S7G-S7K.
See also Figures S2, S4, and S6.
We found that the ciA-B5-bound HNA was still able to interact with anionic liposome and showed pH-dependent conformational change (Figures 3D, S4B, and S7L). This is consistent with the structure showing that the ciA-B5-binding epitope is distant from the “BoNT-switch,” which is a motif crucial for membrane association of HNA at acidic pH (Lam et al., 2018; Figure S4A). However, when we pre-incubated HNA with liposomes to allow membrane insertion of HNA, co-sedimentation of ciA-B5 with liposome-embedded HNA was drastically reduced (Figures 3D and S7M). This finding suggested that ciA-B5 may inhibit HNA from forming a membrane-embedded channel. Previous studies showed that peripherally bound proteins, but not transmembrane or lipid-anchored proteins, could be extracted by either 1 M sodium chloride (pH 4.4), 0.1 M sodium bicarbonate (pH 11), or phosphate-buffered saline (PBS) (pH 7.4) (Bai et al., 2000; Sun et al., 2011). We found that HNA remained bound to liposomes under all these protein extraction conditions, suggesting that HNA stably inserts into the liposome. In contrast, about half of the HNA–ciA-B5 complex was extracted by alkaline solution at pH 11 and by PBS to a less extent, indicating that binding of the HNA–ciA-B5 complex to lipids is partially reversible (Figures 3E and S7N). Taken together, these findings demonstrate that ciA-B5 is able to block membrane insertion of the N-terminal region of HNA and, thus, channel formation of BoNT/A1.
VHHs ciA-H7 and ciA-D12 Interfere with the Unfolding of the Protease Domain
VHHs ciA-H7 and ciA-D12 strongly bind the holotoxin through the surface-exposed regions on LC/A (Mukherjee et al., 2012). The strong neutralizing capacity of VHH ciA-H7 was not readily explained by its structure because its binding site on LC/A is distant from the SNAP-25 binding pocket or the active site of LC/A and has no known function (Figures 4A and S4C). Indeed, ciA-H7 and ciA-D12 did not inhibit cleavage of SNAP-25 by LC/A (residues 2–438) in vitro (Figure S4D). Therefore, the neutralization activity of ciA-H7 seems to be unrelated to the protease function of LC/A. We noticed that ciA-H7 directly interacts with many secondary structures on LC/A through a very large interface (~1,170 Å2), covering residues in the long helix α5, three surface loops (Loop120, 170, and 250) and α10 of LC/A (Figures 4A, S2I, and S2L). In contrast, the binding interface for ciA-D12 is smaller (~778 Å2), mainly including residues in the C-terminal loop, the N-terminal loop, and α10 of LC/A. Interestingly, in our earlier study, we found that another VHH (ciA-F12), which is not protective against BoNT/A1 in mouse studies, recognizes LC/A through an even smaller interface (~684 Å2) involving two surface β-hairpins(Gu et al., 2012; Mukherjee et al., 2012; Figures 4A and S4C). These structural findings not only explain the higher binding affinity of ciA-H7 in comparison to ciA-D12 and ciA-F12 (Mukherjee et al., 2012) but also suggest that ciA-H7 may be superior at stabilizing the structure of LC/A. Because LC must partially unfold upon endosomal acidification before it can pass through the HN channel (~15 Å diameter) to enter the cytoplasm (Koriazova and Montal, 2003), agents preventing LC unfolding in endosomes could prevent translocation and, thus, neutralize the toxin. Alternatively, ciA-H7 might interfere with LC interactions with the membrane and the HN chaperone during translocation and, thus, block trans-membrane delivery of LC/A (Montecucco et al., 1988; Pirazzini et al., 2016; Schiavo et al., 1990).
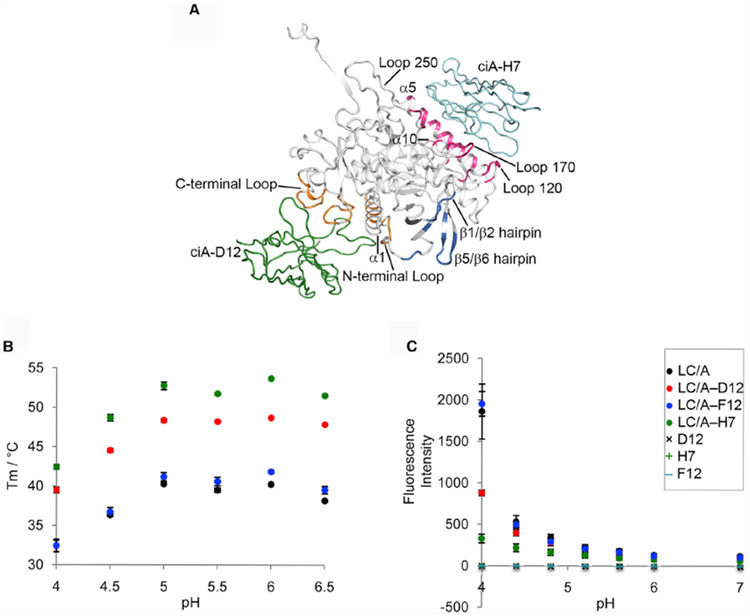
Figure 4. VHH ciA-H7 Interferes with Low-pH-Triggered LC/A Unfolding.
(A) Structure of ciA-D12- and ciA-H7-bound LCHN/A. Only the BoNT LC domain is shown for clarity. The ciA-H7-, ciA-D12-, and ciA-F12-binding epitopes on LC/A are colored pink, orange, and blue, respectively. The corresponding secondary structures are labeled. VHHs ciA-H7 and ciA-D12 are drawn as cyan and green ribbons, respectively.
(B) Thermal stability of LC/A, LC/A–ciA-D12, LC/A–ciA-H7, and LC/A–ciA-F12. The thermal stability of the proteins was measured using a fluorescence-based thermal shift assay on a StepOne real-time PCR system (ThermoFisher). Protein melting was monitored using a hydrophobic dye, SYPRP Orange (Sigma-Aldrich), as the temperature was increased in a linear ramp from 20°C to 90°C. The midpoint of the protein-melting curve (Tm) was determined using software provided by the instrument manufacturer. The data are presented as mean ± SD; n = 3.
(C) ANS fluorescence assay. LC/A (residues 2–438) at 0.5 mg/ml (0.98 mM) was incubated with equimolar ratio of VHH in a buffer containing either 50 mM sodium acetate (pH 4.0–6.0), sodium citrate (pH 6.0), or HEPES (pH 7.0). All buffers contain 100 mM NaCl and 100 mM ANS. The mean values of fluorescence intensity at 478 nm are shown. Error bar represents SD of three replicate experiments.
See also Figures S2, S4, and S6.
We then asked how these VHHs may affect the thermo-stability and low-pH-induced unfolding of LC/A. Using a fluorescence-based thermal shift assay (Figure 4B), we found that the melting temperature (Tm) of LC/A was significantly increased by 30.9%–34.9% at pH 4–6.5 in the presence of ciA-H7 and 20.1%–25.4% for ciA-D12, whereas ciA-F12 had no effect. We further monitored the effect of these VHHs on low-pH-induced unfolding of LC/A by using a hydrophobic dye, ANS. A large increase in fluorescence at pH below 4.8 was indicative of exposure of hydrophobic surfaces during LC/A unfolding. We found that LC/A unfolding was almost completely inhibited by ciA-H7 and by ciA-D12 to a lesser extent but not by ciA-F12 (Figure 4C). Together, these data demonstrate that ciA-H7 is able to potently prevent LC/A unfolding, which is a prerequisite for membrane translocation of LC/A. VHH ciA-D12 has a weaker ability to prevent LC/A unfolding, which may explain why ciA-D12 is a comparatively poor LC/A neutralizing agent in cell-based assays (Mukherjee et al., 2012).
Structure-Based Design of VHH Heterodimers
In earlier studies, we often observed dramatically increased antitoxin potency when we genetically linked two VHHs and produced them as heterodimeric VNAs (Mukherjee et al., 2012). However, these VNAs were designed empirically without knowing the binding sites, neutralizing mechanisms, and their relative positioning of the parental VHHs. Having obtained precise structural information on many high-affinity VHHs against BoNT/A1 and BoNT/B1, we were now able to rationally design a new generation of heterodimeric VNAs, which are composed of two selected VHHs that could bind and/or neutralize toxin synergistically. Specifically, the two parental VHHs in each designer VNAs were carefully selected so that a flexible spacer could be designed based on the distances between their amino or carboxyl termini to allow simultaneous binding to two epitopes on the same toxin molecule. When possible, we paired two VHHs with complementary neutralizing mechanisms. We used the rigid, inflexible portion of VHHs as building blocks, which begin in the framework 1 sequence “QVQLVE…” and end with the framework 4 sequence “…VSS.” All pairwise distance estimates between VHHs were calculated from these termini. The flexible spacers engineered to join two VHHs consisted of either GGGGS repeats or poly-glycines, and their spanning lengths can be accurately estimated (Chen et al., 2013).
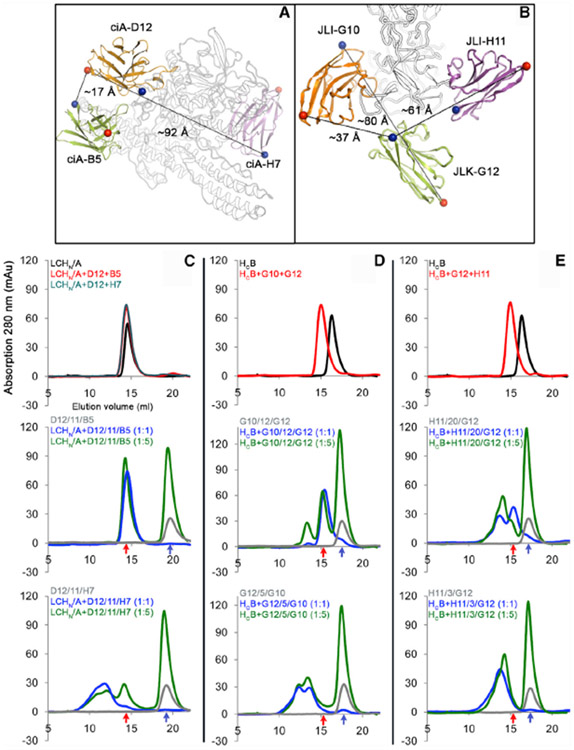
To test this rational design platform, we designed a series of VNAs containing linkers of different lengths that would allow only one VHH to bind at a time or both VHHs to bind simultaneously. In one example, the LC/A-binding ciA-D12, a poorly neutralizing VHH, was found to bind an epitope on BoNT/A1 holotoxin that is spatially proximal to the epitope of ciA-B5, a high-affinity neutralizing VHH binds on HNA. As the estimated distance is ~17 Å and ~20 Å in the ciA-D12/ciA-B5 and ciA-B5/ciA-D12 orientations, respectively (Figure 5A; Table S3), we designed an 11-aa peptide linker composed of glycine and serine to ensure simultaneous binding of ciA-D12 and ciA-B5 (D12/11/B5). Because the binding sites for ciA-D12 and ciA-B5 are close to each other, making it difficult to design a shorter spacer to preclude simultaneous binding, we constructed another VNA as a control, in which ciA-D12 was linked to ciA-H7 that has comparable neutralization potency as ciA-B5 but a binding site distant from ciA-D12 (Mukherjee et al., 2012). These two VHHs were linked with an 11-aa spacer (D12/11/H7), which is too short to permit simultaneous binding.
Figure 5. Rational Design of VNAs to Allow Simultaneous Binding of the Parental VHHs to a Single Toxin Molecule.
(A and B) Inter-molecular distances between selected VHHs that target BoNT/A1 (A) and BoNT/B1 (B), respectively. The Cα atoms of the N- and C-terminal residues are shown in blue and red spheres, respectively.
(C–E) Gel filtration analysis to distinguish simultaneous versus non-simultaneous binding VNAs. D12/11/B5 and D12/11/H7 (C) were examined using LCHN/A. G10/12/G12 and G12/5/G10 (D) and H11/20/G12 and H11/3/G12 (E) were examined using HCB. Elution profiles were shown for the toxin fragments in complex with the parental VHHs (top panel), designer VNAs with optimal spacers to allow simultaneous binding (middle panel), or control VNAs with spacers too short to allow simultaneous binding (bottom panel). The toxin fragments were incubated with parental VHHs at 1:1 molar ratio. The bivalent VNAs were tested at 1:1 (blue curve) or 1:5 (green curve) molar ratio. The elution profiles of the free VNAs are colored gray. The peak elution volumes of the monomeric VNA-toxin complex and the VNA alone are indicated by red and blue arrows, respectively.
See also Figure S4.
The second set of VNAs were engineered based on VHHs JLI-G10 and JLK-G12, which bind at nearby sites on HCB and block BoNT/B1 recognition of its receptors. We engineered two VNAs with these two VHHs in opposite orientations: (1) JLI-G10 was linked to JLK-G12 (distance ~37 Å) with a 12-aa spacer (G10/12/G12) that should permit simultaneous binding, and (2) JLK-G12 was linked to JLI-G10 (distance ~80 Å) with a short 5-aa spacer (G12/5/G10) to prevent simultaneous binding. A third set of VNA was designed based on JLK-G12 and JLI-H11, which is another HCB-binding neutralizing VHH. We designed two heterodimers of these two VHHs in the JLI-H11/JLK-G12 orientation (distance ~61 Å) with either a 20-aa spacer (H11/20/G12) to permit simultaneous binding or a 3-aa spacer (H11/3/G12) that is too short for simultaneous binding (Figure 5B; Table S3).
Because these bifunctional VNAs intrinsically have extremely high affinity for their targets, it is infeasible to differentiate simultaneous from non-simultaneous binding of parental VHHs by SPR-based methods. Therefore, we performed size-exclusion chromatography to examine the binding stoichiometry between BoNT domains and VNAs, as VNAs with simultaneous binders should form a monomeric complex with BoNT, whereas VNAs with non-simultaneous binders should cross-link BoNTs to form high-molecular-weight oligomers (Figure 5). We found that D12/11/B5 and LCHN/A formed monomeric complexes when these proteins were pre-incubated in 1:1 and 5:1 molar ratios (Figure 5C). Simultaneous binding of these two VHHs was further confirmed by the co-crystal structure of LCHN/A in complex with the designer VNA (Figures S4E-S4G). In contrast, the non-simultaneous binding D12/11/H7 cross-linked LCHN/A to form multimeric species with higher molecular weight (Figure 5C, bottom panel). A significant amount of VNAs G10/12/G12 (Figure 5D) and H11/20/G12 (Figure 5E) formed monomeric complexes with HCB, indicating simultaneous binding. But their counterparts G12/5/G10 and H11/3/G12, the predicted non-simultaneous binders, only formed oligomeric complexes due to cross-linking of HCB (Figures 5D and 5E, bottom panels). These data, thus, confirmed that the structure-based design of these VNAs was successful.
Simultaneous Binding VNAs Possess Significantly Improved Antitoxin Potency
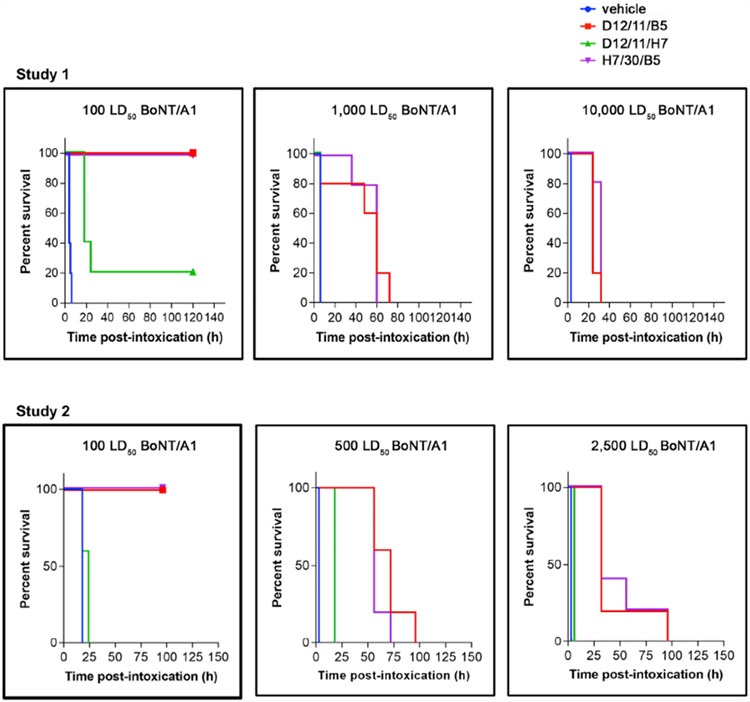
We next compared the in vivo potency of selected VNAs in a mouse co-intoxication model, in which fixed amounts of VNAs were pre-mixed with increasing doses of BoNT/A1 or BoNT/B1 and injected into mice, and the mice were monitored for symptoms of intoxication and time to death (Figures 6 and 7). We first compared the simultaneous binder D12/11/B5 with a non-simultaneous binder, D12/11/H7. VNA D12/11/B5 (~40 pmoles) fully protected mice from 100 lethal dose 50 (LD50) of BoNT/A1 and delayed death for about 2 days in mice receiving 500–1,000 LD50 of toxin, whereas the D12/11/H7-treated mice survived for 1 day when challenged with 100 LD50 and displayed little protection at high challenge doses (Table S3). Only one parental VHH (ciA-B5 or ciA-H7) could neutralize BoNT/A1 in these two VNAs. We previously reported a VNA (H7/30/B5), in which two strong neutralizing VHHs were connected by a 30-aa peptide linker (Mukherjee et al., 2012). But with the new structural information, it is now clear that this peptide linker is too short to allow simultaneous binding of ciA-H7 and ciA-B5. Interestingly, we found that the antitoxin efficacy of D12/11/B5 was comparable to H7/30/B5. This finding suggests that the neutralizing potency of ciA-B5 could be enhanced by a non-neutralizing VHH through synergistic binding in a designer VNA, which could be as potent as a simple addition of two neutralizing VHHs.
Figure 6. BoNT/A1 Intoxication in Mice Is Prevented by Co-administration of the Designer VNAs.
Symptoms of BoNT/A1 intoxication and lethality were monitored following administration of bivalent VNAs (D12/11/B5, D12/11/H7, or H7/30/B5) or toxin only. Time to death is plotted as % survival following co-injection of the indicated dose of BoNT/A1 and VNAs in groups of five mice. See also Figure S5.
Figure 7. BoNT/B1 Intoxication in Mice Is Prevented by Co-administration of the Designer VNAs.
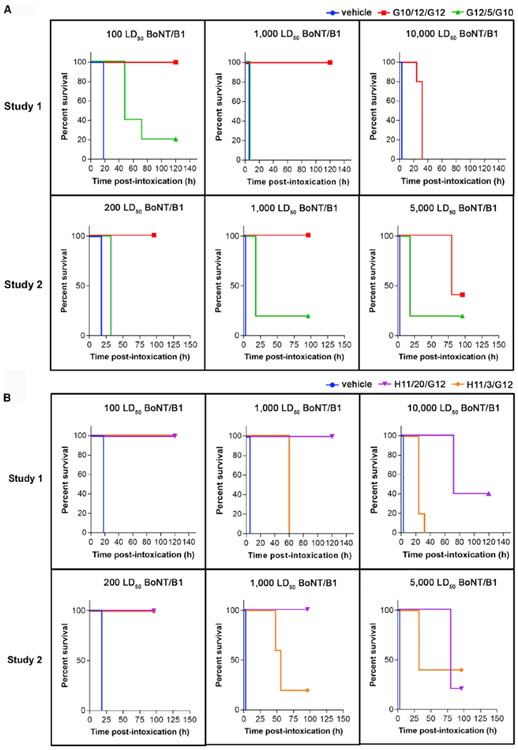
Symptoms of BoNT/B1 intoxication and lethality were monitored following administration of bivalent VNAs G10/12/G12, G12/5/G10(A), H11/20/G12, H11/3/G12 (B), or toxin only. Time to death is plotted as % survival following co-injection of the indicated dose of BoNT/B and VNAs in groups of five mice. See also Figure S5.
We then tested designer VNAs composed of two neutralizing VHHs against BoNT/B1 (Figure 7; Table S3). For designer VNAs H11/20/G12 and G10/12/G12 that allow simultaneous binding of parental VHHs, the VNA-treated mice survived a LD50 challenge with no or minor signs of botulism, respectively. They partially protected mice when exposed to 5,000 LD50 of toxin and significantly delayed death even at 10,000 LD50 of toxin. In contrast, the control VNAs H11/3/G12 and G12/5/G10, which contain the same parental VHHs as the above two, provided little or no protection in mice challenged with 1,000 LD50 of BoNT/B1. We speculate that such dramatically improved antitoxin potency for H11/20/G12 and G10/12/G12 is caused by both synergistic binding and complementary neutralizing mechanisms of the two parental VHHs. Taken together, these results clearly demonstrate the advantage of the structure-based VNA design platform.
We further evaluated the practical application of these VNAs as potential antitoxins by testing the in vivo potency of selected VNAs when they were administered separately from toxins (Figures S5A and S5B). As a proof of concept, VNAs D12/11/B5 or H11/20/G12 were first administered to mice by tail vein injection, which was followed by giving variable doses of BoNT/A1 or BoNT/B1, respectively, by intraperitoneal injection 5 min after VNA administration. We found that both D12/11/B5 and H11/20/G12 fully protected mice from 100 LD50 of the appropriate toxin and delayed death for several days in mice receiving 500–1,000 LD50 of the toxin. These two VNAs fully protected mice against 10 LD50 of BoNT/A1 and BoNT/B1, respectively, when administered 30 or 60 min prior to toxins (Figures S5C and S5D).
DISCUSSION
BoNTs can be extremely toxic to animals and humans, and the source bacteria are widely distributed in nature and express BoNTs with broad sequence variations. There is a clear unmet need for improved treatments for botulism, which should be safe, economical, and easily manufactured in large amounts. In this paper, we strive to develop a platform to rationally design potent VHH-based antitoxins against BoNTs. Specifically, we first investigated the binding epitopes and neutralizing mechanisms for a selected group of VHHs. Based on these structural and functional information, we further enhanced the potency of these VHHs by rational design of heterodimeric VNAs, which use custom-designed peptide linkers to achieve simultaneous binding of two parental VHHs that have non-overlapping yet closely apposed epitopes (Beirnaert et al., 2017; Desmyter et al., 2017).
We revealed two neutralizing mechanisms against BoNT/A1: (1) ciA-B5 blocks the acidic pH-driven insertion of HN into the endosomal membrane, which is required for translocation of LC into the cytosol; and (2) ciA-H7 inhibits the pH-dependent conformational change and translocation of LC. We also reported the creation of a panel of VHHs that bind to BoNT/B1, and we identified three high-affinity, neutralizing VHHs, namely, JLK-G12, JLI-G10, and JLI-H11, which interfere with receptor binding of BoNT/B1. Interestingly, although JLK-G12 and JLI-G10 primarily prevent BoNT/B1 binding to Syt II or ganglioside, respectively, both VHHs also interact with a membrane-binding loop (HC-loop) on HCB. The newly discovered HC-loop is located proximately in between the Syt- and the ganglioside-binding pockets on HCB, serving as the third membrane anchoring point for BoNT/B on neuronal cell surface (Stern et al., 2018). JLK-G12 and JLI-G10 are unique in a way that they can block membrane binding of BoNT/B by targeting the HC-loop. A similar neutralizing mechanism could be applied to antagonize BoNT/G or /DC, as they share similar receptor-recognition mechanisms as BoNT/B (Stern et al., 2018; Zhang et al., 2017). On the other hand, BoNT/A, /HA, and other BoNTs that rely on SV2 as their protein receptor have spatially distant binding sites for SV2 and ganglioside (Stenmark et al., 2008; Yao et al., 2016). It remains unknown whether these BoNTs may also have a membrane-binding motif equivalent to the HC-loop of BoNT/B that could be targeted by antibodies.
Our designer VNAs display significantly improved antitoxin potencies versus component VHHs. In one example, a poorly neutralizing VHH (ciA-D12), when appropriately linked to a neutralizing VHH (ciA-B5) that binds at an adjacent site, resulted in a VNA that is as potent as a simple addition of two neutralizing VHHs. Combining two neutralizing VHHs in a simultaneous binding designer VNA further improved the overall potency, as demonstrated by H11/G12 and G10/G12 neutralization of BoNT/B1. Despite the short half-life of H11/20/G12 and G10/12/G12 in blood (estimated at 1–2 h), they were as potent as a simple combination of two neutralizing mAbs that inhibit BoNT/B dual-receptor binding at non-overlapping epitopes (Chen et al., 2012). Furthermore, we have shown that the potency of VNAs can be further enhanced by including two epitopic tags and co-administering a single anti-tag mAb, which promotes rapid serum clearance of the toxin (Mukherjee et al., 2012). Therefore, designer VNAs hold great promise as anti-BoNT therapeutic agents.
The designer VNA strategy could also help to tackle another major obstacles in the development of botulism antitoxins, which is the high sequence diversity within each BoNT serotypes. For example, BoNT/A and /B each have eight distinct subtypes, with additional subtypes likely to be discovered. It is well known that primary sequence differences among BoNT subtypes can impact antibody binding and neutralization properties (Smith et al., 2005). We found that the ciA-B5 epitope of BoNT/A is identical among BoNT/A1 and A5–A8 and is >90% conserved among BoNT/A2–A4 (Figure S6). Hence, ciA-B5 is likely to bind well to all BoNT/A subtypes. Sequence analysis showed that the JLI-G10, JLK-G12, and JLI-H11 epitopes are moderately conserved among BoNT/B subtypes, suggesting that these VHHs likely have some affinity toward most or all BoNT/B subtypes (Figure S6). Therefore, a comprehensive structure-based mapping of antibody binding epitopes on BoNTs is needed to identify more conserved epitopes on BoNTs, which will aid the development of neutralizing antibodies with cross-subtypes or cross-serotype efficacies.
We expect that designer VNAs will have better tolerance to cross-subtype sequence diversity because they recognize a much larger surface on BoNTs than individual VHHs (Laursen et al., 2018), and the two parental VHHs bind synergistically and could exploit complementary neutralizing mechanisms. In addition, the many co-crystal structures of BoNT–VHH complexes presented here and in the literature lay the foundation for structure-based engineering of VNAs to enhance their neutralizing potencies and broaden reactivity. Taken together, we have established a platform to advance our ability to rapidly and rationally develop novel designer VNAs to prevent and/or treat BoNT intoxication.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to the Lead Contact, Rongsheng Jin (r.jin@uci.edu). All unique/stable reagents generated in this study will be made available on request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Five alpacas, two juveniles (~age 1) and three adults including both males and females, all outbred and raised on pasture, were procured from local farm. All studies were carried out in strict accordance with the recommendations delineated in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The procedures used were approved by the Tufts University Institutional Animal Care and Use Committee (IACUC) and were performed under Protocols #G2011-08 and #G2017-18.
CD1 Mice (eight week old, female) were purchased from Charles River Laboratories. All procedures were approved by the USAMRICD IACUC and conducted in accordance with the principles stated in the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act of 1966 (P.L. 89-544), as amended.
METHOD DETAILS
Cloning, expression, and purification of recombinant proteins
Five alpacas were immunized with purified recombinant catalytically inactive BoNT/B1 holotoxin (ciBoNT/B1) (Mukherjee et al., 2012) and/or LC/B by five multi-site subcutaneous (SC) injections at approximate 3-week intervals essentially as described by Vrentas et al. (2016). Blood was obtained for lymphocyte preparation 3–5 days after the fifth immunization, and RNA was prepared from lymphocytes using the RNeasy kit (QIAGEN, Valencia, CA). Three VHH-display phage libraries were prepared from the lymphocyte RNA from either one alpaca or pools of two alpacas, each as described previously by Moayeri et al. (2015). Three different libraries were prepared, each having a complexity of > 107 independent clones with > 95% containing VHH inserts.
The phage libraries were panned to select and identify VHHs binding to ciBoNT/B1 using methods essentially as previously described (Moayeri et al., 2015; Mukherjee et al., 2012). Some VHHs were obtained by panning on ciBoNT/B1 that had been immobilized either in Nunc Immunotubes or on Costar tissue culture plastic dishes. To select for VHHs binding to native ciBoNT/B1, a further round of panning was performed in which phage from one library was selected for binding to soluble ciBoNT/B1 that had been captured onto Nunc Immunotubes that were first coated with the ciBoNT/B1-binding VHH JEQ-H11 (see Table S1). Two rounds of panning, one at low stringency and a second at higher stringency were performed as described by Vrentas et al. (2016). Typically, at least ninety-five random clones from selected populations obtained using each panning method were screened for expression of VHHs that bound to ciBoNT/B1. A total of 55 VHHs that appeared to be unique based on DNA fingerprinting were selected for DNA sequencing to identify VHHs that derive from common B cell precursors based on homology within the CDRs. From this information, 21 unique VHHs were selected for expression and characterization. Following characterization, 15 clonally independent groups were identified. The Figure S1A shows the sequences of 17 unique selected VHH, including two pair of apparently related VHHs (JLK-G12 and JLO-G11; JLI-G10 and JFM-A11), which derive from different libraries (Table S1). The monomeric VHH coding DNAs were initially expressed and purified as His6-tagged recombinant Escherichia coli thioredoxin (Trx) fusions with a carboxy-terminal E-tag, as previously described (Mukherjee et al., 2014).
LCHN/A (M1–K871) was amplified from catalytically inactive BoNT/A that carries three mutations (E224Q/R363A/Y366F) (Gu et al., 2012) and cloned into pCDF-duet vector following an N-terminal His6 tag and a PreScission cleavage site. Rat Syt II (M1–I61) (Jin et al., 2006), Syt II (M1–C90), LC/A (P2–K438), HCB (E859–E1291) (Jin et al., 2006), ciA-B5, three anti-BoNT/B1 VHHs (JLK-G12, JLI-H11, and JLI-G10), and D12/11/B5 were cloned into pGEX-6P-1 for expression following the N-terminal GST and a PreScission cleavage site. Two anti-BoNT/A1 VHHs (ciA-D12 and ciA-H7) were cloned into pGEX-4T-2 vector following the N-terminal GST and a thrombin cleavage site. To facilitate co-expression of HCB and VHHs, three anti-BoNT/B1 VHHs (JLK-G12, JLI-H11, and JLO-G11) were additionally cloned into pAC28 vector following the N-terminal His6 tag (Kholod and Mustelin, 2001). HNA, HNA S622C/V653C (HNADS), ALc-B8 and ciA-F12 were expressed and purified as described previously (Gu et al., 2012; Lam et al., 2018; Tremblay et al., 2010).
His6-LCHN/Ai was expressed in the E. coli strain BL21-RIL (DE3) (Agilent). Transformed bacteria were grown at 37°C in TB medium in the presence of spectinomycin and chloramphenicol. Expression was induced with 0.1 mM isopropyl-b-D-thiogalactopyranoside (IPTG) when OD600 reached ~1.0. Temperature was then decreased to 25°C and expression was continued for 16 hr. Syt II (1–61), Syt II (1–90), LC/A, HCB, ciA-B5, ciA-D12, ciA-H7, and D12/11/B5 were expressed in E. coli strain BL21-Star (DE3) (Invitrogen). GST-tagged JLK-G12, JLI-H11, and JLI-G10 were expressed in the E. coli strain Origami B (DE3) (Novagen). GST-tagged HCB was co-expressed with His-tagged JLK-G12, JLI-H11, or JLO-G11 in E. coli strain BL21-Star (DE3). Bacteria were cultured at 37°C in LB medium containing ampicillin and/or kanamycin. Temperature was reduced to 18°C when OD600 reached ~0.6. Expression was induced with 1 mM IPTG and continued at 18°C for 16 hr. Cells were harvested by centrifugation and stored at −20°C until use.
For protein purification, bacteria were re-suspended in a buffer containing 50 mM Tris (pH 8.0), 400 mM NaCl, and 0.4 mM PMSF and lysed by sonication. His6–LCHN/A was purified using a Ni-NTA (nitrilotriacetic acid, QIAGEN) affinity column in the same buffer supplemented with 20 mM imidazole and subsequently eluted with 250 mM imidazole. The eluted proteins were pooled followed by His-tag removal using PreScission protease and dialysis at 4°C against a buffer composed of 20 mM sodium acetate (pH 4.3) and 100 mM NaCl. This protein was further purified by MonoS ion-exchange chromatography (GE Healthcare) in a buffer containing 20 mM sodium acetate (pH 4.3) and eluted with a NaCl gradient, followed by Superdex-200 size exclusion chromatography (SEC; GE Healthcare) in 150 mM NaCl and 10 mM HEPES (pH 7.5).
All GST-tagged proteins were purified using Glutathione Sepharose 4B resins (GE Healthcare) in 50 mM Tris (pH 8.0), 400 mM NaCl, and eluted from the resins after on-column cleavage using either PreScission protease (HCB, LC/A, ciA-B5, D12/11/B5, JLK-G12, JLI-H11, and JLI-G10) or thrombin (ciA-H7 and ciA-D12). Uncleaved GST-tagged proteins (Syt II (1–61), Syt II (1–90), and GST-F12) were eluted in the same buffer supplemented with 20 mM reduced glutathione. The proteins were further purified by Superdex-200 Increase or Superdex-75 SEC in 10 mM HEPES (pH 7.4) and 150 mM NaCl. The complexes of HCB with JLK-G12, JLI-H11, or JLO-G11 were purified by Ni-NTA column, separately, followed by GST affinity column, on-column GST cleavage using PreScission protease, and Superdex-200 Increase SEC (150 mM NaCl, 10 mM HEPES, pH 7.4). The HCB–JLI-G10 complex was made by mixing the purified HCB and JLI-G10 at a molar ratio of 1:1.5 for 1 hr on ice, followed by purification using Superdex-200 Increase SEC (10mM HEPES, pH 7.4, and 150 mM NaCl). The LCHN/A–B5–D12–H7 complex or the LCHN/A–D12/11/B5–H7 complex was assembled by mixing the purified LCHN/A with the purified VHHs or VNA at a molar ratio of 1:1.5 for 1 hr on ice. The complexes were further purified by Superdex-200 Increase SEC (150 mM NaCl and 10 mM HEPES, pH 7.4). Each protein was concentrated to ~5 mg/ml using Amicon Ultra centrifugal filters (Millipore) and stored at −80°C until further characterization or crystallization.
All VHH heterodimer VNAs coding DNAs were synthesized (Genscript) and expressed either as cytosolic Trx fusion proteins in E. coli, or as secreted proteins in mammalian cells. Bacterial expression, purification, and quantification were performed as previously described (Mukherjee et al., 2014). VNAs used in the mouse studies reported in this work were expressed by mammalian cells and purified from conditioned media by standard nickel affinity methods (Genscript). The purified proteins were analyzed by SDS-PAGE and western blot and shown to have over 95% purity. Quantification was by Bradford assay with BSA standards.
Pull down assay
To examine the interaction between HCB and rat Syt II (1–61), 20 μg of GST–Syt II (1–61) was immobilized on GST resins as the bait in a buffer containing PBS and 0.01% Tween 20. HCB (300 nM) or HCB pre-incubated with 600 nM of JLI-G10, JLK-G12, or JLI-H11 was pull-down as the preys at 8°C for 1 hr. After washing with the same buffer for three times, samples were boiled with SDS loading dye and analyzed by SDS-PAGE. The pull-down assay between HCB and Syt II (1–90) was performed using the same protocol, except that the buffer contained PBS and 0.5% Triton X-100 in the presence or absence of ~60 μM of GT1b. The pull-down samples were boiled with SDS loading dye supplemented with 4 M Urea.
Liposome co-sedimentation assay
Large unilamellar vesicles (LUV) were prepared as previously described (Lam et al., 2018). Briefly, lipids (1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) and 1-palmitoyl-2-(9,10-dibromostearoyl)phosphatidylcholine (BrPC)) (Avanti Polar Lipid) were dissolved in chloroform while GT1b trisodium salt (Santa Cruz Biotechnology) was dissolved in methanol. The lipids at the indicated molar ratios were mixed and then dried under nitrogen gas and placed under vacuum for overnight. The dried lipids were rehydrated and subjected to 5–10 rounds of freezing and thawing cycles. Liposomes were prepared by extrusion through a 200 nm pore membrane using an Avanti Mini Extruder according to the manufacturer’s instructions.
The HCB–GT1b binding experiment was conducted by mixing 1 μM of HCB or HCB pre-incubated with 2 μM of VHH (JLI-G10, JLK-G12, or JLI-H11) with ~80 μg of liposomes composed of 70/20/10 mol% of BrPC/DOPS/GT1b. The protein–liposome mixture was then incubated in a buffer containing 50 mM NaCl, 20 mM MES (pH 6.5) at room temperature for 30 min followed by spinning progressively at 4,000 × , 9,000 × , and 16,000 × g for 30 min each. Supernatant and pellet were separated and analyzed by SDS-PAGE.
To study the effect of ciA-B5 on HNA–lipid interaction, HNA (25–200 nM) or HNA pre-bound with ciA-B5 at 1:1.5 molar ratio, was incubated with ~45 μg of liposomes composed of 80/20 mol% of BrPC/DOPS in a buffer containing 100 mM NaCl and 50 mM sodium acetate (pH 4.6) at room temperature for 1 hr. In some experiments, the HNA-bound liposomes were further mixed with ciA-B5 for 1 hr and centrifuged.
To further characterize the ciA-B5–HNA-liposome association, the centrifuged pellet of liposomes incubated with 100 nM of HNA or HNA–ciA-B5 complex was re-suspended in a buffer containing either PBS, or 1 M NaCl and 50 mM sodium acetate (pH4.6), or 0.15 M NaCl and 50 mM CAPS (pH 11), or 2% Triton X-100 (Sun et al., 2011), before another round of centrifugation.
Calcein dye release assay
Dried lipid containing 90/10 mol% of asolectin/cholesterol was resuspended in 150 mM NaCl, 20 mM HEPES (pH 7.0), 1 mM EDTA, and 50 mM calcein. Free calcein dye was separated from calcein-entrapped liposomes by desalting (Zeba). Fluorescence was measured on a Spectramax M2e cuvette module with excitation at 493 nm and emission at 525 nm. In the assay, liposomes were diluted in 150 mM NaCl, 20 mM sodium acetate (pH 4.6), 1 mM EDTA to give a final concentration of 0.1 mM and incubated until the fluorescence signal was stable. HNA or HNA pre-incubated with ciA-B5 at a 1:1.5 molar ratio was added at 25–400 nM and the fluorescence intensity was recorded for 10 minutes. The reaction was stopped by adding 0.1% Trion X-100. The percentage of fluorescence change was calculated as (F – Finitial)/(Ffinal – Finitial). The initial rate of calcein dye release was deduced from the slope of the linear part of the curve. The experiments were repeated three times independently.
Membrane depolarization assay
Liposomes composed of 70/20/10 mol% of DOPC/DOPS/cholesterol were prepared in 200 mM NaCl, 1 mM KCl, and 10 mM HEPES (pH 7.0). To create a trans-positive membrane potential (+ 135 mV), liposomes were diluted in 200 mM KCl, 1 mM NaCl, and 10 mM sodium acetate (pH 4.6). Membrane potential was monitored using 6 μM ANS. Valinomycin was added at time 0 s to give a final concentration of 30 nM. At 180 s, 40 nM of HNA pre-incubated with 0–200 nM of ciA-B5 or HNA S622C/V653C (HNADS) was added and the fluorescence intensity at 490 nm was monitored for 6 minutes with excitation at 380 nm. The reaction was stopped by adding 2 μM of gramicidin from Bacillus anerinolyticus (Sigma-Aldrich). The fluorescence change relative to the maximal change in the presence of gramicidin was calculated as (F – Finitial)/(Ffinal – Finitial). The experiments were repeated three times independently.
8-anilinonaphthalene-1-sulfonic acid binding assay
LC/A, LC/A-VHH, or VHH was incubated at ~1 μM with 100 μM ANS for 15 min in either 50 mM sodium acetate (pH 4.0–5.6) or 50 mM Bis-Tris (pH 6–7). All buffers contained 100 mM NaCl. Fluorescence intensity were recorded at 25 °C using a Molecular Devices SpectraMax M2e spectrophotometer with excitation at 370 nm and emission at 478 nm. The fluorescence intensity was corrected by subtraction of background fluorescence from ANS in a buffer without protein. Error bars indicate SD of three replicate measurements.
The binding of ANS to HNA was carried out as previously described (Lam et al., 2018). Briefly, HNA, HNA–ciA-B5, or ciA-B5 was incubated at ~1.34 μM with 100 μM of ANS for 15 min in a buffer containing 100 mM NaCl and either 50 mM sodium acetate (pH 4–5.2) or 50 mM Bis-Tris (pH 6–7). The peak emission intensity at 466 nm was recorded.
Thermal denaturation assay
The thermal stability of LC/A or LC/A–VHH was measured using a fluorescence-based thermal shift assay on a StepOne real-time PCR machine (Life Technologies). LC/A with or without VHH were incubated for 30 min in a buffer containing 0.1 M NaCl and either 50 mM sodium acetate (pH 4–5.5) or 50 mM Bis-Tris (pH 6–6.5). Immediately before the experiment, the protein (1 μM) was mixed with the fluorescent dye SYPRO Orange (Sigma-Aldrich). The samples were heated from 20 °C to 90 °C in a standard ramp rate of 1.5°C/min. The midpoint of the protein-melting curve (Tm) was determined using the analysis software provided by the instrument manufacturer. The experiment was performed in triplicate.
Crystallization
Initial crystallization screens were performed using a Gryphon crystallization robot (Art Robbins Instruments) and high-throughput crystallization screen kits (Hampton Research and QIAGEN). Extensive manual optimizations were performed at 18°C when proteins were mixed with reservoir solution at 1:1 ratio.
The best single crystals of HCB–JLI-G10 were grown by the hanging-drop vapor diffusion method at a protein concentration of 5 mg/ml with a reservoir solution containing 15% PEG 20,000 and 0.1 M MES (pH 6.5).
Crystals of HCB–JLK-G12 were obtained by the hanging-drop vapor diffusion method at a protein concentration of 4 mg/ml with a reservoir solution containing 0.2 M ammonium sulfate, 14% PEG 8,000, and 0.1 M MES (pH 6.0).
Crystals of HCB–JLO-G11 were obtained by the sitting-drop vapor diffusion method at a protein concentration of 5 mg/ml with a reservoir solution containing 20% Ethanol and 0.1 M Tris (pH 8.5).
Crystals of HCB–JLI-H11 were obtained by the sitting-drop vapor diffusion method at a protein concentration of 5 mg/ml with a reservoir solution containing 0.1 M KCl, 15% PEG 5,000 MME, and 0.1 M MES (pH 6.5).
Crystals of LCHN/Ai–B5–D12–H7 were grown by the hanging-drop vapor diffusion method at a protein concentration of 5 mg/ml with a reservoir solution containing 11% PEG 20,000 and 0.1 M sodium cacodylate (pH 6.4). Micro-seeding was necessary to obtain single crystals. Despite extensive efforts, crystallization of other LCHN/Ai–VHHs complexes, such as LCHN/Ai–B5–F12 and LCHN/Ai–B5–F12-H7, and the HNA–ciA-B5 complex was not successful.
Crystals of LCHN/Ai–D12/11/B5–H7 were obtained by the hanging-drop vapor diffusion method at a protein concentration of 3 mg/ml with a reservoir solution containing 13% PEG 20,000 and 0.1 M sodium cacodylate (pH 6.6).
Data collection and structure determination
All crystals were cryoprotected in their original mother liquor supplemented with 20%–25% (v/v) ethylene glycol. The X-ray diffraction data for the crystals of HCB–JLK-G12 and LCHN/Ai–B5–D12–H7 were collected at 100 K at beamline BL9-2 at the Stanford Synchrotron Radiation Lightsource (SSRL). All other data were collected at 100 K at the NE-CAT beamline 24-ID-E, Advanced Photon Source (APS). The data were processed with iMOSFLM (Battye et al., 2011) or XDS as implemented in RAPD (https://github.com/RAPD/RAPD) (Kabsch, 2010). Data collection statistics are summarized in Table S2. Structures of the HCB–JLI-G10, HCB–JLO-G11, HCB–JLK-G12, and HCB–JLI-H11 complexes were determined by molecular replacement using the Phaser software (McCoy et al., 2007) with HCB (PDB: 2NM1) (Jin et al., 2006) and the homology models of JLI-G10, JLO-G11, JLK-G12, or JLI-H11 that were built based on a VHH in PDB 5L21 (Yao et al., 2017) as the search models. Structures of the LCHN/Ai–B5–D12–H7 and LCHN/A–D12/11/B5–H7 complexes were solved by molecular replacement with Phaser using LCHN/A (PDB: 3V0A) (Gu et al., 2012) and the homology models of ciA-B5, ciA-D12, and ciA-H7 as the search models. Manual model building and refinement were performed in COOT (Emsley et al., 2010), PHENIX (Adams et al., 2010), and CCP4 suite (Winn et al., 2011) in an iterative manner. The refinement progress was monitored with the free R value using a 5% randomly selected test set (Brünger, 1992). The structures were validated through MolProbity (Chen et al., 2010) and showed excellent stereochemistry. Structural refinement statistics are listed in Table S2. All structure figures were prepared with PyMol (https://pymol.org/2/).
Gel filtration analysis
Toxin fragment (LCHN/A or HCB) was incubated with various VHHs or VNAs at a toxin:VHH/VNA = 1:1 or 1:5 molar ratio in PBS at 4°C for 1 hr. The protein mixtures were then separated by Superdex-200 SEC in PBS.
SNAP-25 endopeptidase assay
LC/A (2–438) at 1 nM was pre-incubated with 10–100 nM VHH for 15 min and subsequently added to 5 μM of SNAP-25(80–206)-His6 in a buffer containing 20 mM HEPES (pH 7.4), 5 μM MgSO4, 5 μM ZnSO4, and 0.1% Tween20. After incubation at room temperature for 30 min, the reactions were stopped by mixing with SDS-loading dye and boiling for 5 min. The samples were subjected to SDS-PAGE analysis.
Characterization of BoNT/B VHH binding and neutralization properties
VHHs (E-tagged) were tested for binding to the various recombinant BoNT preparations (ciBoNT/B1, HCB, LC/B, etc) by standard ELISAs (Mukherjee et al., 2012) using plates coated with 100 μl of 1 μg/ml of the protein, or by first coating plates with 5 μg/ml of a BoNT-binding VHH (myc tagged) followed by a blocking step and then capturing a 1 ug/ml BoNT preparation. Typically ELISAs started at 125 nM VHH and 1:5 dilutions were performed prior to detection with HRP-goat anti-E-tag (Bethyl). BoNT/B neutralization studies were done similar to those previously reported for VHHs binding to BoNT/A (Mukherjee et al., 2012), but employing 1 nM BoNT/B treatments of primary neurons and detecting VAMP cleavage on western blots as a ratio of VAMP to SNAP25 (1:25,000 rabbit anti-SNAP25 combined with 1:1000 rabbit anti-VAMP (Millipore)) and detected with 1:10,000 HRP-goat anti-rabbit IgG (GE Healthcare). BoNT cleaved VAMP is not recognized by the anti-VAMP antibody, so the assay relies on the ratio of intact VAMP:SNAP25 as comparted to controls.
Mouse toxin lethality assay
Mouse lethality assays were performed similarly as previously described (Mukherjee et al., 2012). Briefly, female CD1 mice 22–27 g each (Charles River Labs) were grouped to minimize inter-group weight variation. VHH and VNA proteins were diluted in PBS at 40 pmol per mouse. BoNT holotoxins were prepared separately in PBS + 0.2% gelatin (Sigma-Aldrich). For co-intoxication assay, VNA and BoNT were combined in a total volume of 250 μl and incubated at room temperature for 30 min prior to administration by intraperitoneal injection. For separated administration experiments, VNA was injected to mice via tail vein at 5, 30, or 60 minutes prior to administration of toxins by intraperitoneal injection. Mice were observed at least twice per day and scored for survival and clinical signs using the following rubric: mild abdominal paradox (score of 1); severe abdominal paradox (2); agonal respiratory pattern (3); lethargy (1); generalized body weakness (2); total body paralysis (lack of righting reflex, 3); and death (16). Progression of toxic signs and time to death were determined for each mouse and averaged within each treatment group.
QUANTIFICATION AND STATISTICAL ANALYSIS
All in vitro assays were performed in triplicate. For pulldown and liposome co-sedimentation assays (Figures 2, 3, and S7), protein band intensities was quantified by software ImageJ and the data were reported as mean ± S.D..
DATA AND CODE AVAILABILITY
Atomic coordinates and structure factors for HCB–JLI-G10, HCB–JLO-G11, HCB–JLK-G12, HCB–JLI-H11, LCHN/Ai–D12–B5–H7, and LCHN/Ai–D12/11/B5–H7 have been deposited in the Protein Data Bank under accession codes 6UHT, 6UL4, 6UFT, 6UC6, 6UI1, 6UL6, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HRP-goat anti-E-tag | Bethyl | Cat#:A190-132P; RRID: AB_345220 |
| Rabbit polyclonal anti-SNAP25 | EMD Millipore | Cat#: AB5871; RRID: AB_2192200 |
| Rabbit polyclonal anti-VAMP | Millipore | Cat#: AB5856; RRID: AB_92090 |
| HRP-goat-anti-rabbit IgG | GE Healthcare | Cat#: NA935; RRID: AB_772207 |
| Bacterial and Virus Strains | ||
| E. coli BL21 Star (DE3) | Invitrogen | Cat#: C6010-03 |
| E. coli BL21-CodonPlus(DE3)-RIL | Agilent Technologies | Cat#: 230245 |
| E. coli Origami B | Novagen | Cat#: 70837-3 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Precision Protease | This Paper | N/A |
| Human alpha-thrombin | Haematologic Technologies | Cat#: HCT-0020 |
| Inactive BoNT/B1 | This Paper | N/A |
| VNA D12/11/B5 (Mukherjee et al., 2012) | Genscript | N/A |
| VNA D12/11/H7 (Mukherjee et al., 2012) | Genscript | N/A |
| VNA H7/30/B5 (Mukherjee et al., 2012) | Genscript | N/A |
| VNA G10/12/G12 (Figure S1) | Genscript | N/A |
| VNA G12/5/G10 (Figure S1) | Genscript | N/A |
| VNA H11/20/G12 (Figure S1) | Genscript | N/A |
| VNA H11/3/G12 (Figure S1) | Genscript | N/A |
| BoNT/A1 | Metabiologics Inc. | Cat#: BoNT/A complex |
| BoNT/B1 | Metabiologics Inc. | Cat#: BoNT/B complex |
| 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) | Avanti Polar Lipid | Cat#: 850375C |
| 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) | Avanti Polar Lipid | Cat#: 840035C |
| 1-palmitoyl-2-(9,10-dibromo)stearoyl phosphatidylcholine (BrPC) | Avanti Polar Lipid | Cat#: 850481C |
| GT1b trisodium salt | Santa Cruz Biotechnology | Cat#: sc-221661 |
| Asolectin | Sigma-Aldrich | Cat#: 11145 |
| Cholesterol | Avanti Polar Lipid | Cat#: 700000 |
| Gramicidin | Sigma Aldrich | Cat#: G5002 |
| SYPRO Orange | Sigma Aldrich | Cat#: S5692 |
| Critical Commercial Assays | ||
| Rneasy Kit | QIAGEN | Cat#: 74104 |
| Crystal Screen HT | Hampton Research | Cat#: HR2-130 |
| SaltRx HT | Hampton Research | Cat#: HR2-136 |
| Index HT | Hampton Research | Cat#: HR2-134 |
| ProComplex Suite | QIAGEN | Cat#: 130915 |
| Deposited Data | ||
| HCB–JLI-G10 structure | PDB | 6UHT |
| HCB–JLK-G12 structure | PDB | 6UFT |
| HCB–JLO-G11 structure | PDB | 6UL4 |
| HCB–JLI-H11 structure | PDB | 6UC6 |
| LCHN/Ai–B5–D12–H7 structure | PDB | 6UI1 |
| LCHN/Ai–D12/11/B5–H7 structure | PDB | 6UL6 |
| Experimental Models: Cell Lines | ||
| Rat primary cerebellar neuron cultures | 5-7d Spraque-Dawley rats | (Eubanks et al., 2010) |
| Experimental Models: Organisms/Strains | ||
| Female CD1 Mouse | Charles River Labs | Cat#: CD1(ICR) |
| Alpacas | Local farms | N/A |
| Oligonucleotides | ||
| LcHN(1–871)_F: attactttcatatcttaaattcaatatagaagtattaataa tttacttaatatattcagtaaatgtagataataatctcagtaaatg | This Paper | N/A |
| LcHN(1–871)_R: catttactgagattattatctacatttactgaatatattaa gtaaattattaatacttctatattgaatttaagatatgaaagtaat | This Paper | N/A |
| SytII(1–90)_F: GGCCGGATCCATGAGAAACATCTTCAAG AGGAACCAG | This Paper | N/A |
| SytII(1–90)_R: GGCCGTCGACTTAGCACTTCTTACAGAT GCAGAAGC | This Paper | N/A |
| LC/A(2–438)_F: ctacttattgtatcctttatctaatgatttctatttagaagtt attatccctcttacacatagcaact | This Paper | N/A |
| LC/A(2–438)_R: agttgctatgtgtaagagggataataacttctaaatag aaatcattagataaaggatacaataagtag | This Paper | N/A |
| Recombinant DNA | ||
| pCDF-duet-1 | Novagen | Cat#: 71340 |
| pGEX-6p-1 | GE Healthcare | Cat#: 28954648 |
| pGEX-4t-2 | GE Healthcare | Cat#: 28954550 |
| Software and Algorithms | ||
| iMOSFLM | (Battye et al., 2011) | https://www.mrc-lmb.cam.ac.uk/mosflm/imosflm/ver730/introduction.html |
| XDS | (Kabsch, 2010) | https://github.com/RAPD/RAPD |
| Phaser | (McCoy et al., 2007) | https://www.phenix-online.org/ |
| Coot | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | (Adams et al., 2010) | http://www.phenix-online.org |
| CCP4 | (Winn et al., 2011) | http://www.ccp4.ac.uk/ |
| Pymol | The PyMOL Molecular Graphics System, Version 1.7.Schrödinger, LLC. | https://pymol.org/2/ |
| Clustal Omega | (Sievers et al., 2011) | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| ESPript 3.0 | (Robert and Gouet, 2014) | http://espript.ibcp.fr/ESPript/ESPript/ |
| PDBe-PISA | (Krissinel and Henrick, 2007) | https://www.ebi.ac.uk/pdbe/pisa/ |
| Graphpad v7.2 | Prism | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
Crystal structures of six VHHs that bind and neutralize BoNT/A1 or BoNT/B1
Three VHHs inhibit the membrane insertion or the light chain unfolding of BoNT/A1
Three VHHs block the synaptotagmin, ganglioside, and/or lipid binding of BoNT/B1
Rationally designed bifunctional VHH heterodimers show superior antitoxin potency
ACKNOWLEDGMENTS
The authors acknowledge the excellent technical assistance of Michelle Debatis and the early mouse challenge studies done by Dr. Jean Mukherjee that helped to stimulate these studies. This work was partly supported by National Institute of Allergy and Infectious Diseases (NIAID) grants R01AI139087, R21AI139690, and R21AI123920 to R.J.; R01AI125704 to R.J. and C.B.S.; and Defense Threat Reduction Agency—Chemical Biological Defense Therapeutics funding to P.M.M. NE-CAT at the Advanced Photon Source (APS) is supported by a grant from the National Institute of General Medical Sciences (P41 GM103403). Use of the APS, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under contract no. DE-AC02-06CH11357. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.01.107.
REFERENCES
- Adams PD, Afonine PV, Bunkióczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R, Schmidt JJ, Stafford RG, and Swaminathan S (2009). Mode of VAMP substrate recognition and inhibition of Clostridium botulinum neurotoxin F. Nat. Struct. Mol. Biol 16, 789–794. [DOI] [PubMed] [Google Scholar]
- Bai J, Earles CA, Lewis JL, and Chapman ER (2000). Membrane-embedded synaptotagmin penetrates cis or trans target membranes and clusters via a novel mechanism. J. Biol. Chem 275, 25427–25435. [DOI] [PubMed] [Google Scholar]
- Battye TGG, Kontogiannis L, Johnson O, Powell HR, and Leslie AGW (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr 67, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirnaert E, Desmyter A, Spinelli S, Lauwereys M, Aarden L, Dreier T, Loris R, Silence K, Pollet C, Cambillau C, and de Haard H (2017). Bivalent llama single-domain antibody fragments against tumor necrosis factor have picomolar potencies due to intramolecular interactions. Front. Immunol 8, 867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntsson RP-A, Peng L, Dong M, and Stenmark P (2013). Structure of dual receptor binding to botulinum neurotoxin B. Nat. Commun 4, 2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbach MA, and Brunger AT (2004). Substrate recognition strategy for botulinum neurotoxin serotype A. Nature 432, 925–929. [DOI] [PubMed] [Google Scholar]
- Brünger AT (1992). Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. (2019). Botulism Annual Summary, 2017 (U.S. Department of Health and Human Services, CDC; ). [Google Scholar]
- Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, and Stevens RC (2006). Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature 444, 1096–1100. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang S, Wang H, Mao X, Zhang T, Ji G, Shi X, Xia T, Lu W, Zhang D, et al. (2012). Potent neutralization of botulinum neurotoxin/B by synergistic action of antibodies recognizing protein and ganglioside receptor binding domain. PLoS One 7, e43845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zaro JL, and Shen WC (2013). Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev 65, 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A, Silva DA, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam KH, Yao G, et al. (2017). Massively parallel de novo protein design for targeted therapeutics. Nature 550, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JO, Sherwood LJ, Collazo MT, Garza JA, and Hayhurst A (2010). Llama single domain antibodies specific for the 7 botulinum neurotoxin serotypes as heptaplex immunoreagents. PLoS One 5, e8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmyter A, Spinelli S, Boutton C, Saunders M, Blachetot C, de Haard H, Denecker G, Van Roy M, Cambillau C, and Rommelaere H (2017). Neutralization of human interleukin 23 by multivalent nanobodies explained by the structure of cytokine–nanobody complex. Front. Immunol 8, 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, and Chapman ER (2003). Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J. Cell Biol 162, 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Thompson AA, Fan Y, Lou J, Conrad F, Ho M, Pires-Alves M, Wilson BA, Stevens RC, and Marks JD (2010). A single-domain llama antibody potently inhibits the enzymatic activity of botulinum neurotoxin by binding to the non-catalytic α-exosite binding region. J. Mol. Biol 397, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza Y, Wong D, Ahene A, Der K, Martinez Z, Pham J, Cobb RR, Farr-Jones S, Marks JD, and Tomic MT (2019). Pharmacokinetics of human recombinant anti-botulinum toxin antibodies in rats. Toxins (Basel) 11, E345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubanks LM, Silhár P, Salzameda NT, Zakhari JS, Xiaochuan F, Barbieri JT, Shoemaker CB, Hixon MS, and Janda KD (2010). Identification of a natural product antagonist against the botulinum neurotoxin light chain protease. ACS Med. Chem. Lett 1, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sambashivan S, Brunger AT, and Montal M (2012). Beltless translocation domain of botulinum neurotoxin A embodies a minimum ion-conductive channel. J. Biol. Chem 287, 1657–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A, Ramirez-Franco J, Desplantes R, Debreux K, Ferracci G, Wernert F, Blanchard M-P, Maulet Y, Youssouf F, Sangiardi M, et al. (2019). Gangliosides interact with synaptotagmin to form the high-affinity receptor complex for botulinum neurotoxin B. Proc. Natl. Acad. Sci. USA 116, 18098–18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godakova SA, Noskov AN, Vinogradova ID, Ugriumova GA, Solovyev AI, Esmagambetov IB, Tukhvatulin AI, Logunov DY, Naroditsky BS, Shcheblyakov DV, and Gintsburg AL (2019). Camelid VHHs fused to human fc fragments provide long term protection against botulinum neurotoxin a in mice. Toxins (Basel) 11, E464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Rumpel S, Zhou J, Strotmeier J, Bigalke H, Perry K, Shoemaker CB, Rummel A, and Jin R (2012). Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science 335, 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera C, Tremblay JM, Shoemaker CB, and Mantis NJ (2015). Mechanisms of ricin toxin neutralization revealed through engineered homodimeric and heterodimeric camelid antibodies. J. Biol. Chem 290, 27880–27889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Rummel A, Binz T, and Brunger AT (2006). Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature 444, 1092–1095. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholod N, and Mustelin T (2001). Novel vectors for co-expression of two proteins in E. coli. Biotechniques 31, 322–323, 326–328. [DOI] [PubMed] [Google Scholar]
- Koriazova LK, and Montal M (2003). Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat. Struct. Biol 10, 13–18. [DOI] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Lam KH, Yao G, and Jin R (2015). Diverse binding modes, same goal: The receptor recognition mechanism of botulinum neurotoxin. Prog. Biophys. Mol. Biol 117, 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KH, Guo Z, Krez N, Matsui T, Perry K, Weisemann J, Rummel A, Bowen ME, and Jin R (2018). A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1. Nat. Commun 9, 5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen NS, Friesen RHE, Zhu X, Jongeneelen M, Blokland S, Vermond J, Van Eijgen A, Tang C, Van Diepen H, Obmolova G, et al. (2018). Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 362, 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield MJ, Wentz TG, Zhang S, Lee EJ, Dong M, Sharma SK, and Doxey AC (2019). Bioinformatic discovery of a toxin family in Chryseobacterium piperi with sequence similarity to botulinum neurotoxins. Sci. Rep 9, 1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Leysath CE, Tremblay JM, Vrentas C, Crown D, Leppla SH, and Shoemaker CB (2015). A heterodimer of a VHH (variable domains of camelid heavy chain-only) antibody that inhibits anthrax toxin cell binding linked to a VHH antibody that blocks oligomer formation is highly protective in an anthrax spore challenge model. J. Biol. Chem 290, 6584–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal M (2009). Translocation of botulinum neurotoxin light chain protease by the heavy chain protein-conducting channel. Toxicon 54, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C (1986). How do tetanus and botulinum toxins bind to neuronal membranes? Trends Biochem. Sci 11, 314–317. [Google Scholar]
- Montecucco C, Schiavo G, Gao Z, Bauerlein E, Boquet P, and Das-Gupta BR (1988). Interaction of botulinum and tetanus toxins with the lipid bilayer surface. Biochem. J 251, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J, Tremblay JM, Leysath CE, Ofori K, Baldwin K, Feng X, Bedenice D, Webb RP, Wright PM, Smith LA, et al. (2012). A novel strategy for development of recombinant antitoxin therapeutics tested in a mouse botulism model. PLoS One 7, e29941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J, Dmitriev I, Debatis M, Tremblay JM, Beamer G, Kashentseva EA, Curiel DT, and Shoemaker CB (2014). Prolonged prophylactic protection from botulism with a single adenovirus treatment promoting serum expression of a VHH-based antitoxin protein. PLoS One 9, e106422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak SU, Griffiss JM, McKenzie R, Fuchs EJ, Jurao RA, An AT, Ahene A, Tomic M, Hendrix CW, and Zenilman JM (2014). Safety and pharmacokinetics of XOMA 3AB, a novel mixture of three monoclonal antibodies against botulinum toxin A. Antimicrob. Agents Chemother 58, 5047–5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzini M, Tehran DA, Leka O, Zanetti G, Rossetto O, and Montecucco C (2016). On the translocation of botulinum and tetanus neurotoxins across the membrane of acidic intracellular compartments. Biochim. Biophys. Acta 1858, 467–474. [DOI] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel A (2013). Double receptor anchorage of botulinum neurotoxins accounts for their exquisite neurospecificity. Curr. Top. Microbiol. Immunol 364, 61–90. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Boquet P, Dasgupta BR, and Montecucco C (1990). Membrane interactions of tetanus and botulinum neurotoxins: a photolabelling study with photoactivatable phospholipids. J. Physiol. (Paris) 84, 180–187. [PubMed] [Google Scholar]
- Schmidt DJ, Beamer G, Tremblay JM, Steele JA, Kim HB, Wang Y, Debatis M, Sun X, Kashentseva EA, Dmitriev IP, et al. (2016).A tetraspecific VHH-based neutralizing antibody modifies disease outcome in three animal models of clostridium difficile infection. Clin. Vaccine Immunol 23, 774–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schussler E, Sobel J, Hsu J, Yu P, Meaney-Delman D, Grammer LC 3rd, and Nowak-Wȩgrzyn A (2017). Workgroup Report by the Joint Task Force Involving American Academy of Allergy, Asthma & Immunology (AAAAI); Food Allergy, Anaphylaxis, Dermatology and Drug Allergy (FADDA) (Adverse Reactions to Foods Committee and Adverse Reactions to Drugs, Biologicals, and Latex Committee); and the Centers for Disease Control and Prevention Botulism Clinical Treatment Guidelines Workgroup-Allergic Reactions to Botulinum Antitoxin: A Systematic Review. Clin. Infect. Dis 66, S65–S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheoran AS, Dmitriev IP, Kashentseva EA, Cohen O, Mukherjee J, Debatis M, Shearer J, Tremblay JM, Beamer G, Curiel DT, et al. (2015). Adenovirus vector expressing Stx1/Stx2-neutralizing agent protects piglets infected with Escherichia coli O157:H7 against fatal systemic intoxication. Infect. Immun 83, 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Lou J, Geren IN, Forsyth CM, Tsai R, Laporte SL, Tepp WH, Bradshaw M, Johnson EA, Smith LA, and Marks JD (2005). Sequence variation within botulinum neurotoxin serotypes impacts antibody binding and neutralization. Infect. Immun 73, 5450–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Dupuy J, Imamura A, Kiso M, and Stevens RC (2008). Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 4, e1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D, Weisemann J, Le Blanc A, von Berg L, Mahrhold S, Piesker J, Laue M, Luppa PB, Dorner MB, Dorner BG, and Rummel A (2018). A lipid-binding loop of botulinum neurotoxin serotypes B, DC and G is an essential feature to confer their exquisite potency. PLoS Pathog. 14, e1007048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Suresh S, Liu H, Tepp WH, Johnson EA, Edwardson JM, and Chapman ER (2011). Receptor binding enables botulinum neurotoxin B to sense low pH for translocation channel assembly. Cell Host Microbe 10, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehran DA, and Pirazzini M (2018). Novel botulinum neurotoxins: exploring underneath the iceberg tip. Toxins (Basel) 10, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanongsaksrikul J, Srimanote P, Maneewatch S, Choowongkomon K, Tapchaisri P, Makino S, Kurazono H, and Chaicumpa W (2010). A V H H that neutralizes the zinc metalloproteinase activity of botulinum neurotoxin type A. J. Biol. Chem 285, 9657–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay JM, Kuo CL, Abeijon C, Sepulveda J, Oyler G, Hu X, Jin MM, and Shoemaker CB (2010). Camelid single domain antibodies (VHHs) as neuronal cell intrabody binding agents and inhibitors of Clostridium botulinum neurotoxin (BoNT) proteases. Toxicon 56, 990–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance DJ, Tremblay JM, Mantis NJ, and Shoemaker CB (2013). Stepwise engineering of heterodimeric single domain camelid VHH antibodies that passively protect mice from ricin toxin. J. Biol. Chem 288, 36538–36547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Moayeri M, Keefer AB, Greaney AJ, Tremblay J, O’Mard D, Leppla SH, and Shoemaker CB (2016). A diverse set of single-domain antibodies (VHHs) against the anthrax toxin lethal and edema factors provides a basis for construction of a bispecific agent that protects against anthrax infection. J. Biol. Chem 291, 21596–21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Schmidt D, Liu W, Li S, Shi L, Sheng J, Chen K, Yu H,Tremblay JM, Chen X, et al. (2014). A novel multivalent, single-domain antibody targeting TcdA and TcdB prevents fulminant Clostridium difficile infection in mice. J. Infect. Dis 210, 964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Zhang S, Mahrhold S, Lam KH, Stern D, Bagramyan K, Perry K, Kalkum M, Rummel A, Dong M, and Jin R (2016). N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A. Nat. Struct. Mol. Biol 23, 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Lam KH, Weisemann J, Peng L, Krez N, Perry K, Shoemaker CB, Dong M, Rummel A, and Jin R (2017). A camelid single-domain antibody neutralizes botulinum neurotoxin A by blocking host receptor binding. Sci. Rep 7, 7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Berntsson RP-A, Tepp WH, Tao L, Johnson EA, Stenmark P, and Dong M (2017). Structural basis for the unique ganglioside and cell membrane recognition mechanism of botulinum neurotoxin DC. Nat. Commun 8, 1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for HCB–JLI-G10, HCB–JLO-G11, HCB–JLK-G12, HCB–JLI-H11, LCHN/Ai–D12–B5–H7, and LCHN/Ai–D12/11/B5–H7 have been deposited in the Protein Data Bank under accession codes 6UHT, 6UL4, 6UFT, 6UC6, 6UI1, 6UL6, respectively.