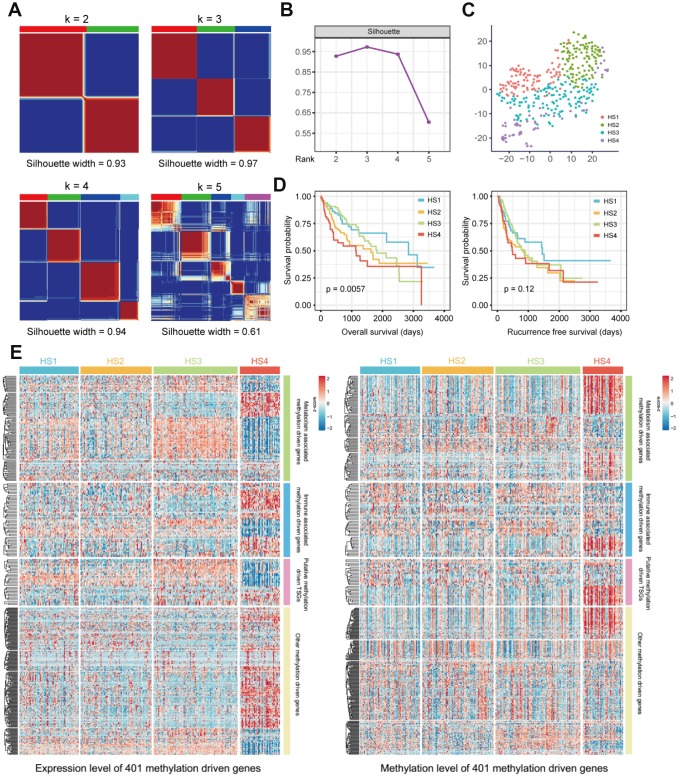
Figure 1.
Identification of HCC subclasses based on integrated transcriptome and methylation data of MDGs. (A) Consensus matrix for k = 2 to k = 5. (B) Silhouette values under corresponding k values. (C) T-SNE analysis of mRNA expression data from tumor samples included in the cluster analysis (D) OS and RFS of 4 HCC subclasses. Statistical significance of differences was determined by Log-rank test. (E) Heatmaps show the expression and methylation level of 401 MDGs in HCC subclasses. 401 MDGs were divided into 4 groups, including metabolism associated MDGs, immune associated MDGs, putative methylation driven TSGs and other MDGs. HCC: hepatocellular carcinoma; MDG: methylation driven gene; t-SNE: t-distributed stochastic neighbor embedding; OS: overall survival; RFS: recurrence free survival; TSG: tumor suppressor genes.