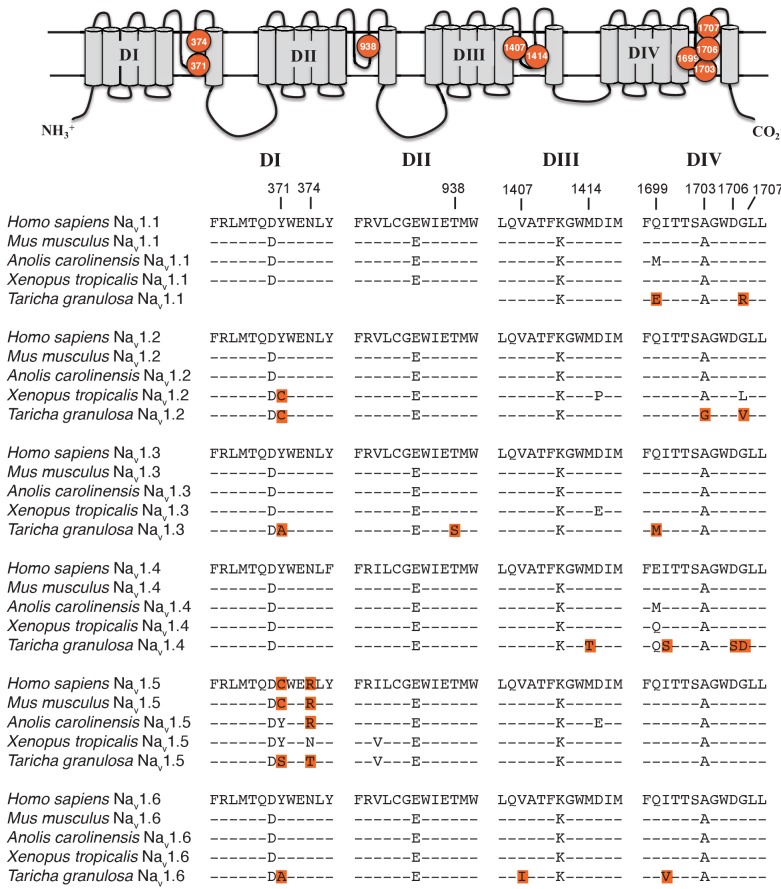
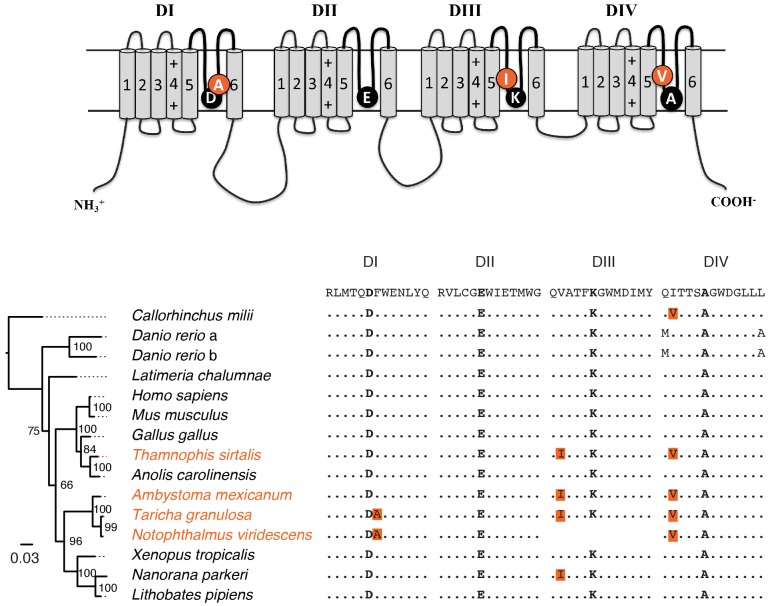
Figure 4. Protein alignment of Nav channels across representative vertebrates.
Sequence alignment of S5-S6 P-loops from newts and other vertebrates showing amino acid substitutions relative to the P-loop consensus sequence for each Nav channel shown here. Putative TTX resistance mutations are highlighted in orange; mutations that are not highlighted are either synapomorphic in a gene clade or are present in TTX sensitive channels. Data are missing for DI and DII of Nav1.1 in newts, which we did not recover in our sequencing efforts. The approximate locations of newt mutations are shown as orange circles, and the amino acid site of each mutation is numbered based on Nav1.6 from Mus musculus.