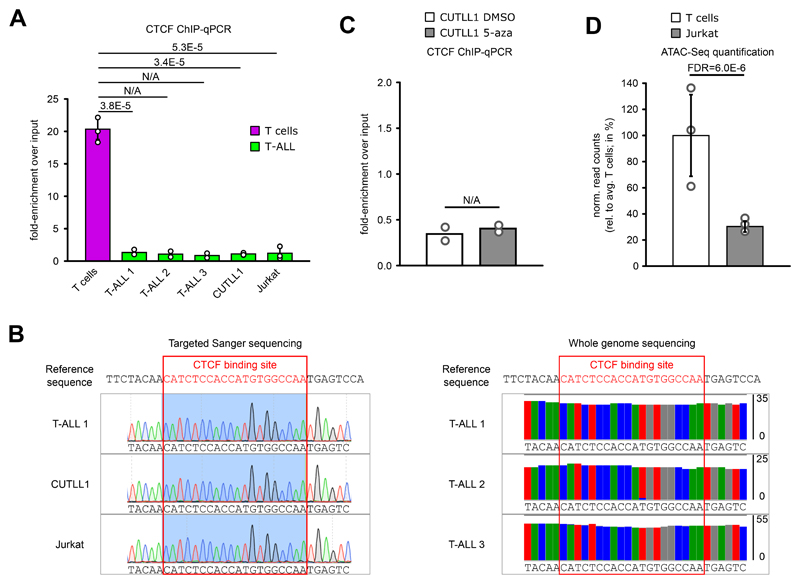
Extended Data Fig. 5. Difference in CTCF insulation in MYC locus is not due to genomic mutation but potentially regulated by open chromatin.
A) CTCF ChIP-qPCR of the CTCF binding site in the lost MYC TAD boundary, shown as fold-enrichment over input. Significant differences compared to T cells were calculated with an unpaired one-sided t test, following the hypothesis of loss of CTCF binding in T-ALL samples as determined from the genome-wide analysis (n = 3 replicates for T cells, T-ALL 1, T-ALL 2, CUTLL1 and Jurkat; n = 2 replicates for T-ALL 3 and T-ALL 4). Error bars indicate s.d.; center value indicates mean.
B) Targeted sanger sequencing indicates no mutation in T-ALL in the CTCF binding site at the MYC TAD boundary. Tracks show chromatogram of individual base calls (left). Whole genome sequencing indicates no mutation in T-ALL in the motif of CTCF binding site. Tracks show (mis-)matches compared to reference sequence in all reads covering the respective genomic position (right).
C) CTCF ChIP-qPCR before and after treatment with global DNA-demethylation agent 5-azacytidine (n = 2 replicates).
D) ATAC-seq quantification for T cells and Jurkat for the genomic area covering loss of CTCF binding in the downstream TAD boundary of MYC. Data was normalized to the average T cell signal, shown in percent (n = 3 replicates). Statistical evaluation was performed using DiffBind with edgeR-method, following multiple testing correction. Error bars indicate s.d.; center value indicates mean.