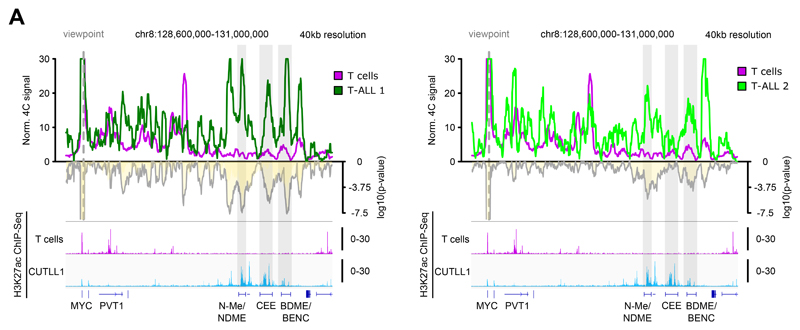
Extended Data Fig. 6. 4C-Seq validation of MYC super-enhancer interaction in primary T-ALL.
A) 4C-seq analysis using MYC promoter as viewpoint. Positive y-axis shows interactions with the MYC promoter viewpoint as normalized read counts, negative y-axis shows significance of differential interactions between T cells and primary T-ALL samples as log10(P value) derived using edgeR function glmQLFTest. H3K27ac ChIP-seq tracks for T cells and CUTLL1 are represented below as fold-enrichment over input. Number replicates: T cells 4C n = 2; T-ALL 1 4C n = 1; T-ALL 2 4C n = 2; T cells H3K27ac n = 2; CUTLL1 H3K27ac n = 2.