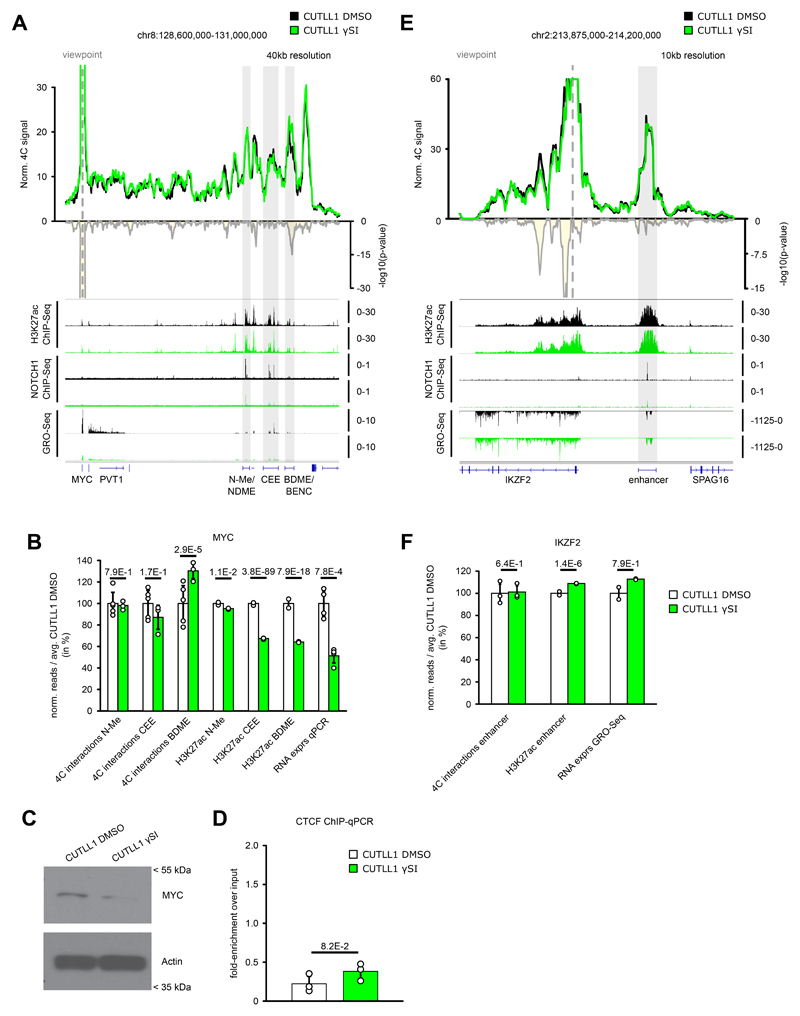
Extended Data Fig. 9. Treatment with γSI does not alter all NOTCH1 dynamic enhancers.
A) 4C-seq using MYC promoter as viewpoint. Positive y-axis shows interactions with viewpoint as normalized read counts, negative y-axis shows significance of differential interactions as log10(P value) calculated using edgeR function glmQLFTest (CUTLL1 DMSO n = 5; CUTLL1 γSI n = 3). Tracks below show H3K27ac, NOTCH1 ChIP-seq and GRO-seq (positive strand only) as fold-enrichment where applicable, and counts-per-million reads otherwise.
B) Quantification of H3K27ac signal (enrichment over input), chromatin interactions by 4C-seq for the interactions of MYC promoter and MYC expression Interaction changes are measured by centering the 40kb bin on highest peaks within N-Me/NDME, CEE or BDME/BENC elements. MYC expression was measured by qPCR. All quantifications are normalized to CUTLL1 DMSO, shown in percent. Error bars indicate s.d.; center value indicates mean. Significance is shown as false-discovery rate (FDR) for H3K72ac signal change (R package DiffBind with edgeR-method), P value for chromatin interaction change (edgeR function glmQLFTest) or one-tailored t test for qPCR changes.
C) Cropped western blot images immunoblotted with MYC antibody. Unprocessed western blots can be found as Source Data. Experiment was repeated twice with similar results.
D) CTCF ChIP-qPCR of lost MYC boundary upon γSI in CUTLL1 (n = 3). Error bars indicate s.d.; center value indicates mean. Significance was calculated using unpaired two-sided t test.
E) 4C-seq analysis using IKZF2 promoter as viewpoint after γSI treatment. Positive y-axis shows normalized read counts, negative y-axis shows significance of differential interactions as log10(P value) calculated using edgeR function glmQLFTest (CUTLL1 DMSO n = 3 ; CUTLL1 γSI n = 3). Tracks below show H3K27ac, NOTCH1 ChIP-seq and GRO-seq (negative strand only) as fold-enrichment over input where applicable, and counts-per-million reads otherwise.
F) H3K27ac signal is specific for enhancer highlighted in D). Interaction changes are measured by centering the 40kb bin on the highest enhancer peak. IKZF2 expression after γSI treatment was measured by GRO-seq. All quantifications are normalized to the average T cell signal, shown in percent. Error bars indicate s.d.; center value indicates mean. Significance is shown as false-discovery rate (FDR) for H3K72ac signal (R package DiffBind with edgeR-method), P value for chromatin interaction (edgeR function glmQLFTest) or one-tailored t test for qPCR expression.