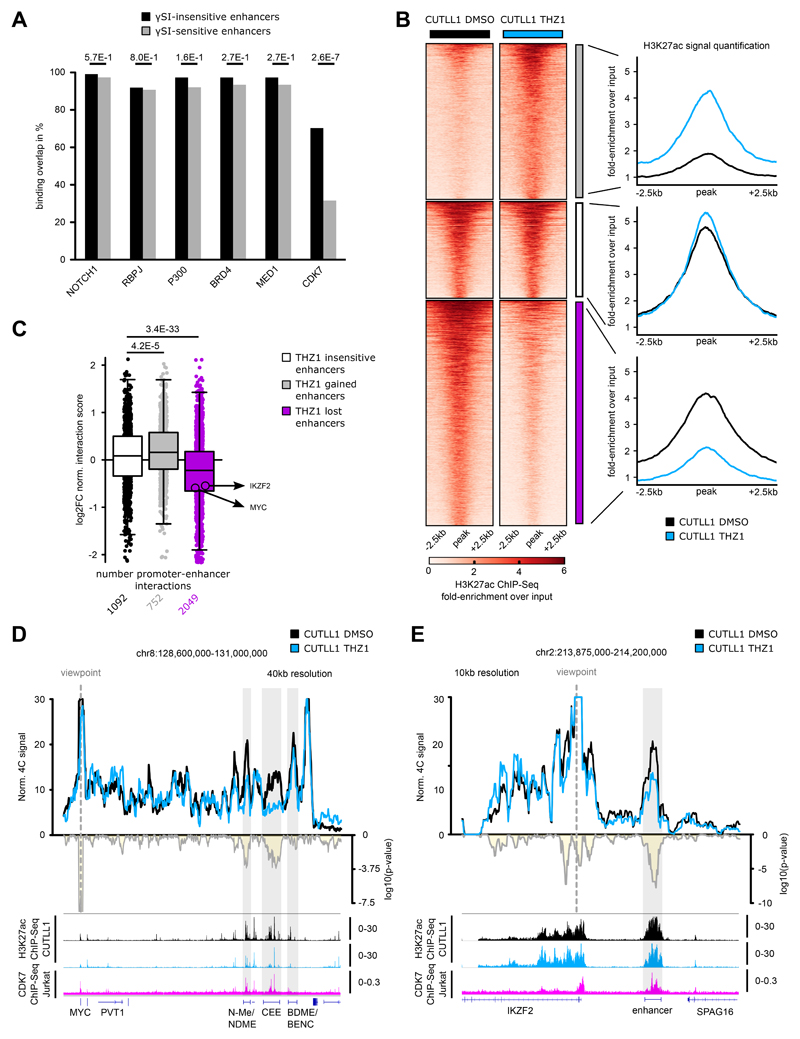
Fig. 6. CDK7 inhibition concomitantly reduces H3K27ac levels and associated promoter-enhancer looping.
A) LOLA analysis for public ChIP-seq data in CUTLL1/Jurkat from the LOLA database with γSI-insensitive and γSI-sensitive enhancers. Statistical differences in overlap between γSI-insensitive and sensitive enhancers with ChIP-seq peaks were calculated using a two-sided Fisher exact test.
B) H3K27ac occupancy in CUTLL1. Groups consist of stable (middle, white, n = 1,396), increased (upper, grey, n = 2,246) and reduced non-promoter H3K27ac signal (lower, pink, n = 3248). Heatmap shows the H3K27ac signal as fold-enrichment over input and line plots depict quantification of H3K27ac signal. Differential analysis was performed with the R package DiffBind with edgeR-method and differential peaks were selected using FDR < 0.05, log2 fold-change > 1.0 or < -1.0 (Number replicates: CUTLL1 DMSO n = 4; CUTLL1 n = 2).
C) Changes in Hi-C interactions between non-promoter H3K27ac peaks defined in B) and connected gene promoters (defined using CUTLL1 H3K27ac HiChIP) are shown as log2 fold-change (average of n = 2 replicates). Each dot represents a promoter-enhancer interaction. Significance of shifts compared to enhancer-promoter interactions associated with stable enhancers is calculated by an unpaired one-sided t test.
D+E) 4C-seq using MYC (D) or IKZF2 promoter (E) as viewpoint. Positive y-axis shows interactions with the viewpoint as normalized read counts, negative y-axis shows significance of differential interactions as log10(P value) calculated using edgeR function glmQLFTest. Tracks below show H3K27ac and CDK7 ChIP-seq track, and represent fold-enrichment over input where applicable and counts-per-million reads otherwise. Number replicates: CUTLL1 DMSO 4C MYC n = 3; CUTLL1 THZ1 4C MYC n = 3; CUTLL1 DMSO 4C IKZF2 n = 3; CUTLL1 THZ1 4C IKZF2 n = 3; CUTLL1 DMSO H3K27ac n = 2; CUTLL1 THZ1 H3K27ac n = 2; Jurkat CDK7 n = 1.