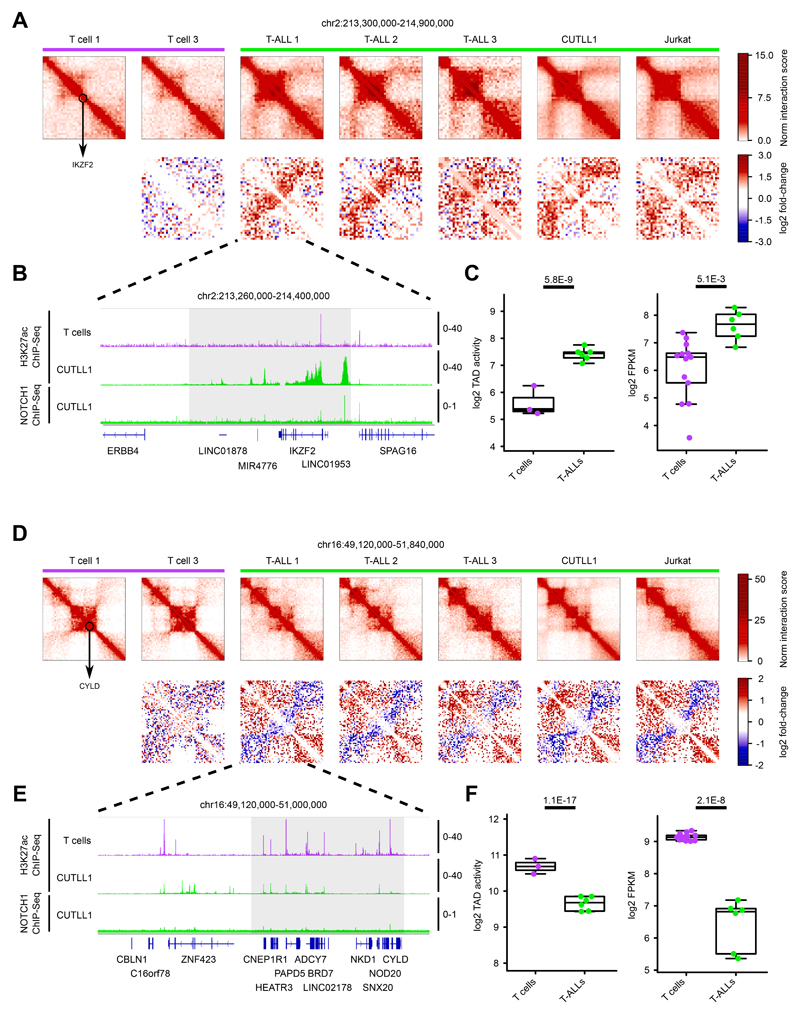
Extended Data Fig. 2. Genomic loci displaying differential intra-TAD activity in T-ALL.
A) Hi-C interaction heatmaps (first row) showing the IKZF2 locus (black circle). Second row shows heatmaps of log2 fold-change interactions compared to T cell 1.
B) H3K27ac ChIP-seq tracks for IKZF2 locus in T cells and CUTLL1, NOTCH1 ChIP-seq tracks for CUTLL1. Tracks represent fold-enrichment over input where applicable and counts-per-million reads otherwise. Number replicates: T cells H3K27ac n = 2; CUTLL1 H3K27ac n = 2; CUTLL1 NOTCH1 n = 1.
C) Quantifications for intra-TAD activity (left; as highlighted in A)) and expression of IKZF2 (right). Statistical evaluation for intra-TAD activity was performed using paired two-sided t test of average per interaction-bin for IKZF2 TAD between T cells (n = 3) and T-ALL (n = 6), followed by multiple testing correction. Log2 FPKM of IKZF2 expression for T cells (n = 13) and T-ALL (n = 6) samples; statistical evaluation was performed using edgeR followed by multiple testing correction.
D) Hi-C interaction heatmaps (first row) showing the CYLD locus (black circle). Second row shows heatmaps of log2 fold-change interactions when compared to T-cell 1.
E) H3K27ac ChIP-seq tracks for CYLD locus in T cells and CUTLL1, NOTCH1 ChIP-seq tracks for CUTLL1. Tracks represent fold-enrichment over input where applicable and counts-per-million reads otherwise. Number replicates: T cells H3K27ac n = 2; CUTLL1 H3K27ac n = 2; CUTLL1 NOTCH1 n = 1.
F) Quantifications for intra-TAD activity (left; as highlighted in D)) and expression of CYLD (right). Statistical evaluation for intra-TAD activity was performed using paired two-sided t test of average per interaction-bin for CYLD TAD between T cells (n = 3) and T-ALL (n = 6), followed by multiple testing correction (see methods). Log2 FPKM of CYLD expression for T cells (n = 13) and T-ALL (n = 6); statistical evaluation was performed using edgeR followed by multiple testing correction.