Fig. 2. Phylogenetic relationship of the Reps from FgGMTV1 and selected circular ssDNA viruses.
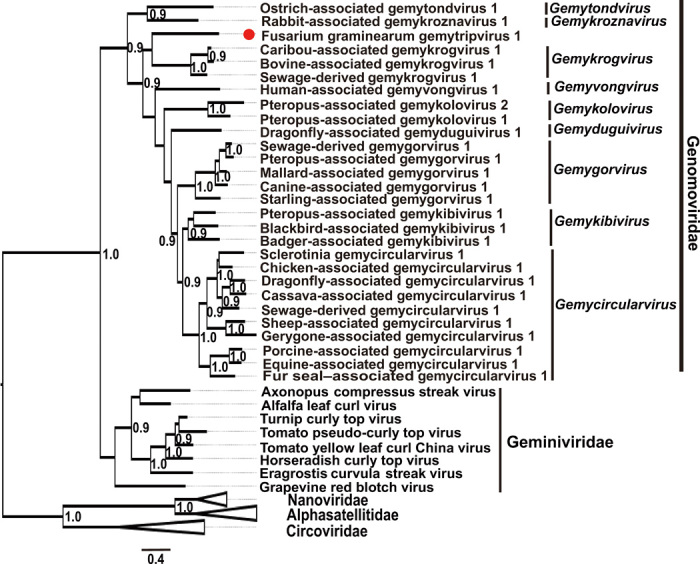
An unrooted phylogenetic tree was constructed by a maximum-likelihood method based on multiple amino acid sequence alignments of the Rep using PhyML 3.0. Ambiguously aligned regions were removed using the online software Gap Strip/Squeeze v2.1.0. The best-fit model “RtREV + G + I + F” was selected using the Smart Model Selection technique implemented in the PhyML 3.0. Numbers at the nodes represent approximate likelihood ratio test values derived using an SH-like calculation (only values greater than 0.9 are shown). The position of FgGMTV1 is indicated by a red dot.
