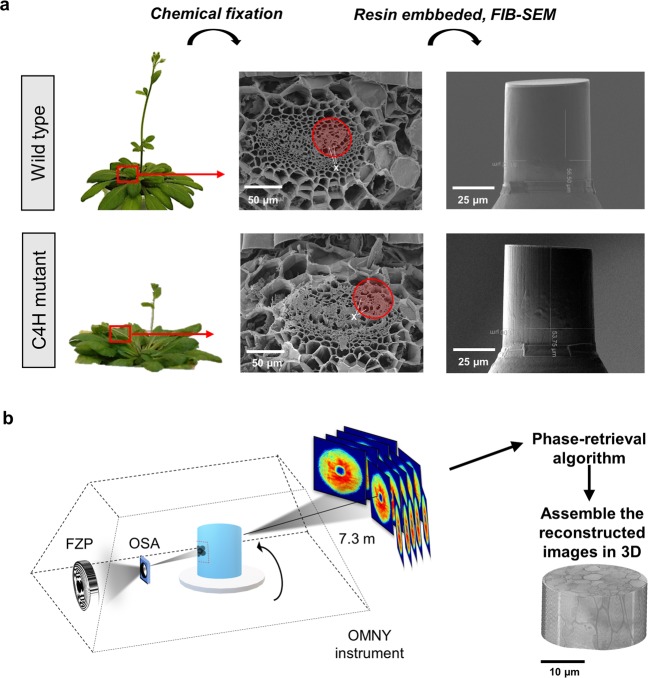
Figure 1.
PXCT experiment workflow. (a) The petioles of wild type and C4H mutant plants were selected (red square), and the overall petiole structure was measured by scanning electron microscopy to identify regions containing vessels (X). The tissue fragments were submitted to chemical fixation, and resin infusion and the 50 µm diameter x 50 µm height regions of interest (red circle) were cut out using FIB-SEM. (b) OMNY instrument used in the cSAXS beamline (PSI) for the X-ray experiment. The sample was inserted into the OMNY cryogenic device34, which uses a Fresnel zone plate (FZP) to focus and define the coherent beam size. A combination of a central stop (CS) and an order sorting aperture (OSA) are used as filters, such that only the first diffraction order makes it to the sample. This removes additional diffraction orders as well as the undiffracted component of the beam. In the ptychographic measurements, the sample was scanned with overlapping coherent X-ray illumination for each tomographic projection between 0 and 180°. Each two-dimensional diffraction pattern is recorded in the far-field and the phase is retrieved by iterative algorithms that reconstruct a real-space two-dimensional projection. The set of two-dimensional projections are then used to reconstruct the tomographic image.