Abstract
Recently, the long noncoding RNA (lncRNA) plasmacytoma variant translocation 1 (PVT1) was reported to be involved in the pathogenesis of several cancers, including human colorectal cancer (CRC). However, the molecular basis for cancer initiation, development, and progression remains unclear. In this study, we observe that upregulated PVT1 is associated with poor prognosis and bad clinicopathological features of CRC patients. In vitro means of PVT1 loss in a CRC cell line inhibit cell proliferation, migration, and invasion. Furthermore, dual-luciferase reporter and RNA pull-down assays indicated that PVT1 binds to miR-16-5p, which has been shown to play strong tumor suppressive roles in CRC. Targeted loss of miR-16-5p partially rescues the suppressive effect induced by PVT1 knockdown. Vascular endothelial growth factor A (VEGFA), a direct downstream target of miR-16-5p, was suppressed by PVT1 knockdown in CRC cells. Overexpression of VEGFA is known to modulate the AKT signaling cascade by activating vascular endothelial growth factor receptor 1 (VEGFR1). We, therefore, show that PVT1 loss combined with miR-16-5p overexpression reduces tumor volume maximally when propagated within a mouse xenograft model. We conclude that the PVT1-miR-16-5p/VEGFA/VEGFR1/AKT axis directly coordinates the response in CRC pathogenesis and suggest PVT1 as a novel target for potential CRC therapy.
Keywords: PVT1, miR-16-5p, VEGFA, AKT, CRC
Introduction
Colorectal cancer (CRC) is the third most common cancer among men and women in the United States.1 The World Health Organization (WHO) European Region (ER) reported 471,000 new cases each year and a mean mortality rate of 28.2 per 100,000 population.2 Despite progress for marker identification used in early diagnosis, prognosis, and prediction of CRC patients’ response to treatment in the past, most of the molecular biomarkers remain generally inconsistent for accurate diagnosis and patient prognosis. Therefore, we address an unmet need to discover new potential biomarkers for prognosis, prediction, and deeper elucidation of the exact molecular mechanisms underlying CRC.
Noncoding RNAs longer than 200 nt are typically classified as long noncoding RNAs (lncRNAs) and have been shown to actively participate in regulation of targeted gene expression at epigenetic, transcriptional, and posttranscriptional levels.3 lncRNA plasmacytoma variant translocation 1 (PVT1), located at 8q24.21, is upregulated in many cancers and associated with cell proliferation, apoptosis, lymph node invasion and metastasis, and tumor prognosis.4, 5, 6, 7, 8 A previous study9 has shown that PVT1 knockdown could enhance radiosensitivity of non-small-cell lung cancer (NSCLC) cells partly through inhibiting cell proliferation and inducing apoptosis by sponging miR-195. An additional study by Kong et al.10 identified that PVT1 played a strong oncogenic role in gastric cancer partly through epigenetic regulation of p15 and p16. Moreover, PVT1 is also considered to be associated with epithelial-mesenchymal transition (EMT) in esophageal cancer,11 pancreatic cancer,12,13 ovarian cancer,14 and breast cancer.15 In recent years, several reports have revealed that PVT1 is related to the pathogenesis of CRC;16,17 however, its precise molecular mechanism for this cancer type remains unclear.
MicroRNAs (miRNAs) are a group of short noncoding RNAs 21–23 nt long that have been demonstrated to be involved in tumorigenesis.18,19 Emerging analysis has demonstrated that miRNAs can be detected in the serum of CRC patients with fidelity and represent a potent signature for CRC. Furthermore, a study by Chen et al.20 detected 69 miRNAs in the serum of CRC patients, and among these miR-485-5p, miR-361-3p, miR-326, and miR-487b were found to be unique in CRC patients. Recently, new miRNAs have emerged that predict the occurrence of CRC with greater accuracy, such as miR-195-5p,21 miR-215,22 as well as some others not described herein.
Increasing evidence has indicated that lncRNAs can serve as competing endogenous RNAs (ceRNAs) during oncogenesis and can function as miRNA sponges, thereby suppressing the regulatory effect of miRNAs on mRNAs.23, 24, 25, 26 However, the regulatory roles of lncRNA PVT1 to act as “miRNA sponges” in CRC remain largely unknown. In our study, using starBase (http://starbase.sysu.edu.cn/), HMDD v2.0 (http://www.cuilab.cn/hmdd), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), we identified PVT1 as a strong candidate target of miR-16-5p.
Based on these initial characterizations, we confirm the role of PVT1 and miR-16-5p in directing CRC progression as a principal underlying mechanism for human CRC.
Results
lncRNA PVT1 Is Significantly Upregulated in CRC and Associated with CRC Progression and Poor Prognosis
To elucidate the role of PVT1 in CRC, we first tested the expression level of PVT1 in 72 pairs of CRC tissues, and four CRC cell lines were analyzed by quantitative real-time reverse transcription PCR (RT-PCR). We observed that expression levels of PVT1 were upregulated in most CRC specimens (Figure 1A) and cell lines (Figure 1B) compared with matched normal tissues and the epithelial cell line FHC, respectively (p < 0.05). Using a median PVT1 value as a cutoff, among the 72 CRC patients, 47 cases were PVT1 high and 25 cases were considered low in PVT1 (PVT1 low). Further statistical analyses showed that increased expression of PVT1 was significantly associated with lymph node metastasis (p < 0.01), distant metastasis (p < 0.05), and TNM (tumor, node, metastasis) stage (p < 0.05) (Table 1). Moreover, CRC patients with high levels of PVT1 reflected significantly shorter overall survival than did those with low expression of PVT1 by the analysis of a Kaplan-Meier survival curve (p < 0.05) (Figure 1C). Next, the receiver operating characteristic (ROC) curve was performed to assess the sensitivity and specificity of PVT1 expression in differentiating CRC from normal tissues (cutoff value = 1.045, Youden index = 0.597, sensitivity = 0.653, specificity = 0.944). Notably, PVT1 showed remarkable predictive significance with an area under the ROC curve of 0.81 (p < 0.01) (Figure 1D). Thus, elevated expression of PVT1 was significantly associated with poor prognosis for CRC patients.
Figure 1.
lncRNA PVT1 Is Significantly Upregulated in CRC and Associated with CRC Progression and Poor Prognosis
(A and B) PVT1 expression was validated by quantitative real-time RT-PCR in CRC tumor samples (A) and cell lines (B). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments. (C) Kaplan-Meier analysis of correlation between the PVT1 levels and overall survival of CRC patients. (D) ROC curve analysis for the prediction of CRC using quantitative real-time RT-PCR-detected PVT1 expression.
Table 1.
Correlations between Long Noncoding RNA PVT1 Expression and Clinical Characteristics in CRC Patients (n = 72)
| Patient Characteristics | Number | PVT1 Lowa | PVT1 Higha | p Value | χ2 |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 39 | 14 (36%) | 25 (64%) | 0.82 | 0.052 |
| Female | 33 | 11 (33%) | 22 (67%) | ||
| Age (years) | |||||
| <6 | 25 | 9 (36%) | 16 (64%) | 0.86 | 0.028 |
| ≥60 | 47 | 16 (34%) | 31 (66%) | ||
| Tumor size (cm) | |||||
| < | 33 | 13 (40%) | 20 (60%) | 0.44 | 0.587 |
| ≥5 | 39 | 12 (31%) | 27 (69%) | ||
| Histological differentiation | |||||
| Well | 6 | 2 (33%) | 4 (67%) | 0.821 | 0.395 |
| Moderate | 58 | 21 (36%) | 37 (64%) | ||
| Poor | 8 | 2 (25%) | 6 (75%) | ||
| Lymph node metastasis | |||||
| N0 | 33 | 18 (55%) | 15 (45%) | 0.001∗∗ | 10.56 |
| N1–N2 | 39 | 7 (18%) | 32 (82%) | ||
| Distant metastasis | |||||
| M0 | 47 | 21 (45%) | 26 (55%) | 0.015∗ | 5.923 |
| M1 | 25 | 4 (16%) | 21 (84%) | ||
| TNM stage | |||||
| I | 2 | 2 (100%) | 0 (0%) | 0.02∗ | 9.75 |
| II | 6 | 4 (67%) | 2 (33%) | ||
| III | 58 | 19 (33%) | 39 (67%) | ||
| IV | 6 | 0 (0%) | 6 (100%) | ||
| CEA (ng/mL) | |||||
| < | 51 | 18 (35%) | 33 (65%) | 0.87 | 0.025 |
| ≥5 | 21 | 7 (33%) | 14 (67%) |
∗p < 0.05, ∗∗p < 0.01.
Using median PVT1 values as the cutoff.
Knockdown of PVT1 Inhibits the Malignant Characteristics of CRC Cell Lines
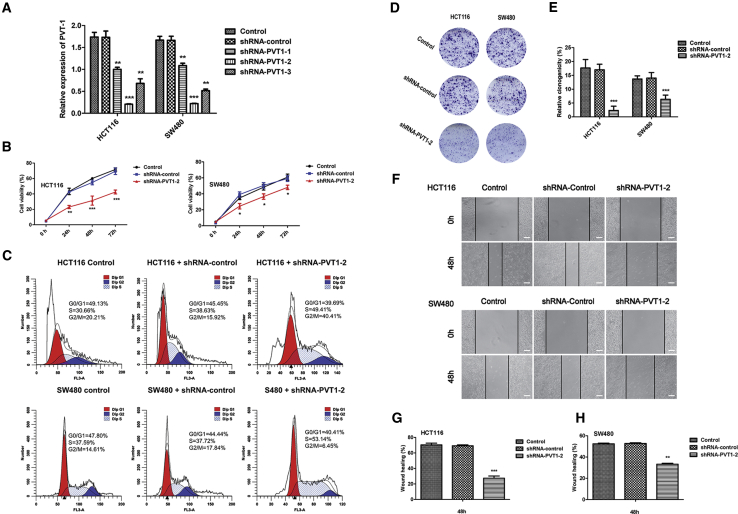
In order to investigate the oncogenic function roles of PVT1 in CRC, we designed three short hairpin RNAS (shRNAs) to silence PVT1 lncRNA expression in HCT116 and SW480 cell lines, and these shRNAs decreased PVT1 expression at different levels; we then targeted PVT1 using shRNA-PVT1-2 for the subsequent experiments due to its highest inhibitory efficiency (Figure 2A). Cell proliferation assays with a Cell Counting Kit-8 (CCK-8) indicated that silencing of PVT1 corresponds with the loss of cell proliferation in two cell lines (Figure 2B). Then, we performed fluorescence-activated cell sorting (FACS) to detect cellular DNA content changes during the cell cycle. We show that PVT1 knockdown in both the HCT116 and SW480 cell lines displayed a significant increase in the percentage of cells in the S phase (Figure 2C). The colony formation (Figures 2D and 2E) and scratch wound healing assays (Figures 2F–2H) showed that PVT1 knockdown significantly suppresses invasion and migration in HCT116 and SW480 cell lines.
Figure 2.
Knockdown of PVT1 Inhibits the Malignant Characteristics of CRC Cell Lines
(A) The interfering efficacies of three PVT1-targeting shRNAs on PVT1 were measured by quantitative real-time RT-PCR. ∗∗p < 0.01, ∗∗∗p < 0.001 versus shRNA-control. (B) Graph depicting cell proliferation rates by CCK-8 assay. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus shRNA-control. (C) FACS analysis shows the phases of the cell cycle and the percentage of cells in each cell cycle. (D and E) Colony formation assay showed shRNA-PVT1-2 significantly inhibited the cells colony formation both in HCT116 and SW480 cells. ∗∗∗p < 0.001 versus shRNA-control. (F–H) Scractch wound healing assays showed shRNA-PVT1-2 significantly inhibited the cells migration abilities both in HCT116 and SW480 cells. ∗∗p < 0.01, ∗∗∗p < 0.001 versus shRNA-control. Scare bars, 100 μm. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments.
lncRNA PVT1 Acts as a miRNA Sponge and Negatively Regulates miR-16-5p Expression
Increasing evidence demonstrates that lncRNAs could serve as miRNA sponges to regulate target gene expression via competition with endogenous RNAs. Using starBase (http://starbase.sysu.edu.cn/), HMDD v2.0 (http://www.cuilab.cn/hmdd), and miRTarBase (http://mirtarbase.mbc.nctu.edu.twphp/index.php), we identified miR-15a-5p, miR-15b-5p, miR-16-5p, miR-17-5p, and miR-195-5p as candidates that bind the PVT1 lncRNA and correspond with outcomes in human CRC. However, among these miRNAs, miR-16-5p has been demonstrated to be closely related to the pathogenesis of CRC through both in vitro and in vivo experiments.27, 28, 29 Considering that the relationship between miR-16-5p and PVT1 remains unclear, we therefore focused on the specific mechanism of PVT1-miR-16-5p in the pathogenesis of CRC. The potential binding sites of PVT1 with miR-16-5p were predicted using starBase (Figure 3A). Luciferase reporter assays demonstrated that overexpression of miR-16-5p significantly decreased the luciferase activity of the vector containing the complete PVT1 sequence (p < 0.01) but did not affect the luciferase activity of the vector with the mutant miR-16-5p-binding site (Figure 3B). To further demonstrate the direct binding of miR-16-5p and PVT1, we utilized biotin-labeled miR-16-5p and its mutant mimics to pull down PVT1 in HCT116 and SW480 cells with PVT1 overexpression, and the results showed that wild-type (WT) miR-16-5p captured more PVT1 compared with the mutant (Figure 3C). We then evaluated the expression of miR-16-5p in 72 pairs of CRC specimens. Quantitative real-time RT-PCR results showed that miR-16-5p was significantly downregulated (p < 0.05) (Figure 3D). Moreover, PVT1 expression was negatively correlated with the expression of miR-16-5p in CRC tissues (r = −0.321, p < 0.01) (Figure 3E). As expected, silencing of PVT1 increased the miR-16-5p expression in HCT116 and SW480 cell lines (Figure 3F). Taken together, these data demonstrated that PVT1 acts as a miRNA sponge for miR-16-5p in CRC.
Figure 3.
lncRNA PVT1 Acts as a miRNA Sponge and Negatively Regulates miR-16-5p Expression
(A) The putative miR-16-5p binding sites with the PVT1 sequence 3′ UTR are shown. (B) Luciferase reporter gene assays were used to evaluate the interaction between miR-16-5p and PVT1. ∗∗p < 0.01 versus the PVT1-WT + miR-NC group. (C) The biotinylated wild-type and mutant miR-16-5p were transfected into HCT116 and SW480 cells, respectively. The expression level of PVT1 was tested by quantitative real-time RT-PCR after streptavidin capture. ∗∗∗p < 0.001 versus biotin-miR-16-5p-WT. (D) miR-16-5p expression was validated by quantitative real-time RT-PCR in CRC tumor samples. ∗p < 0.05. (E) Relationship between PVT1 and miR-16-5p expression in CRC tissues. p < 0.01. (F) miR-16-5p expression was validated by quantitative real-time RT-PCR in CRC cell lines transfected with shRNA-control or three PVT1-targeting shRNAs. ∗p < 0.05 versus shRNA-control. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments.
Overexpression of miR-16-5p Inhibits the Malignant Characteristics of CRC Cell Lines
HCT116 and SW480 cells were transfected with miR-16-5p agomir (agomir-16-5p) or miR-16-5p antagomir (antagomir-16-5p) to upregulate or downregulate miR-16-5p, respectively. As in Figures 4A and 4B, compared with the agomir-NC group, the proliferation was inhibited in the agomir-16-5p group (p < 0.05). Additionally, the agomir-16-5p caused cells to stay in the S phase (Figure 4C). Furthermore, invasion (Figures 4D and 4E) and migration (Figures 4F–4H) were also inhibited in the agomir-16-5p group (p < 0.05). Meanwhile, the abilities of proliferation, invasion, and migration were enhanced in the antagomir-16-5p group (Figures 4A–4H) (p < 0.05).
Figure 4.
Overexpression of miR-16-5p Inhibits the Malignant Characteristics of CRC Cell Lines
(A and B) Effects of miR-16-5p on proliferation of HCT116 (A) and SW480 (B) cells. ∗p < 0.05, ∗∗p < 0.01 versus control + agomir-NC; #p < 0.05, ##p < 0.01 versus control + antagomir-NC. (C) FACS analysis showing the phases of the cell cycle and the percentage of CRC cells in each cell cycle. (D and E) Colony formation assay showed agomir-16 significantly inhibited the cells colony formation, but the antagomir-16's effect is adverse. ∗∗∗p < 0.001 versus control + agomir-NC; #p < 0.05 versus control + antagomir-NC. (F–H) Scratch wound healing assays showed that agomir-16 significantly inhibited the cells migration, however the antagomir-16's effect is adverse. ∗p < 0.05, ∗∗p < 0.01 versus control + agomir-NC; #p < 0.05 versus control + antagomir-NC. Scare bars, 100 μm. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments.
The Suppressive Effects on CRC Cells Induced by PVT1 Knockdown Are Mediated by miR-16-5p
After the luciferase reporter and RNA pull-down assays, PVT1 was confirmed to be a target of miR-16-5p. Whether miR-16-5p was involved in the inhibitory effects on CRC cells induced by PVT1 knockdown needed to be clarified. The proliferation (Figures 5A and 5B), invasion (Figures 5D–5F), and migration (Figures 5G–5I) assays showed that CRC cells cotransfected with short hairpin (sh)-PVT1 and agomir-16-5p exhibited stronger inhibitory effects than did the cells cotransfected with sh-NC (negative control) and agomir-NC (p < 0.05). Additionally the proportion of cells cotransfected with sh-PVT1 and agomir-16-5p in the S phase were the highest (Figure 5C). Conversely, antagomir-16-5p partially reversed the tumor suppressive effects induced by sh-PVT1 (Figures 5A–5I). The cells cotransfected with sh-RNA and agomir or antagomir were all observed in a laser confocal microscope (Figure S1), and our results showed that the agomir-16-5p and antagomir-16-5p had no effect on PVT1 expression (Figure S2).
Figure 5.
The Suppressive Effects on CRC Cells Induced by PVT1 Knockdown Are Mediated by miR-16-5p
(A and B) CCK-8 assay to evaluate the effects of PVT1 combined with miR-16-5p on cell proliferation of HCT116 (A) and SW480 (B) cells. ∗∗p < 0.01 versus control; ###p < 0.001 versus control + sh-NC + agomir-NC. (C) FACS analysis to evaluate the effects of PVT1 combined with miR-16-5p on cell cycle and the percentage of CRC cells in each cell cycle. (D–F) Colony formation assay showed sh-PVT1-2 inhibited the cells colony formation, and the agomir-16 with a synergy effect. ∗∗∗p < 0.001 versus control; ###p < 0.001 versus control + sh-NC + agomir-NC. (G–I) Scratch wound healing assays showed sh-PVT1-2 inhibited the cells migration, and the agomir-16 with a synergy effect. ∗p < 0.05, ∗∗p < 0.01 versus control; ##p < 0.01, ###p < 0.001 versus control + sh-NC + agomir-NC. Scare bars, 100 μm. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments.
lncRNA PVT1 Knockdown Suppresses CRC Progression via Inhibiting miR-16-5p-Mediated VEGFA/VEGFR1/AKT Signaling
Overwhelming evidence had demonstrated that vascular endothelial growth factor A (VEGFA) could bind to vascular endothelial growth factor receptor 1 (VEGFR1) and then activate AKT, which plays key roles in cancer proliferation, metastasis, angiogenesis, and survival.30, 31, 32 We initially measured VEGFA and phosphorylated (p-)AKT protein levels in SW480 and HCT116 cells. These results show that VEGFA and p-AKT were significantly upregulated in SW480 and HCT116 cells (Figure 6A).
Figure 6.
lncRNA PVT1 Knockdown Suppresses CRC progression via Inhibiting miR-16-5p-Mediated VEGFA/VEGFR1/AKT Signaling
(A) Western blot analysis of the protein expression of VEGFRA and phosphorylated AKT in FHC, HCT116, and SW480 cells. ∗∗p < 0.01, ∗∗∗p < 0.001 versus FHC. (B and C) VEGFA and phosphorylated AKT, ERK, and JNK protein expression in PVT1 knockdown of HCT116 (B) and SW480 (C) cells. ∗∗∗p < 0.001 versus shRNA-control. (D and E) Effects of miR-16-5p on the VEGFA and phosphorylated VEGFR1, AKT, and eNOS protein expression in HCT116 (D) and SW480 (E) cells. ∗∗p < 0.01, ∗∗∗p < 0.001 versus control + agomir-NC; #p < 0.05 versus control + antagomir-NC. (F and G) VEGFA and phosphorylated VEGFR1, AKT, and eNOS protein expression in HCT116 (F) and SW480 (G) cells with or without VEGFA overexpression or silence. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus control + VEGFA(+)-NC; #p < 0.05, ###p < 0.001 versus control + VEGFA(−)-NC. (H and I) Effects of PVT1 combined with miR-16-5p on VEGFA and phosphorylated VEGFR1, AKT, and eNOS protein expression in HCT116 (H) and SW480 (I) cells. ∗∗∗p < 0.001 versus control + sh-NC + agomir-NC; #p < 0.05, ##p < 0.01, ###p < 0.001 versus control + sh-PVT1-2. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments.
To further demonstrate whether PVT1 knockdown inhibited CRC development through the VEGFA-mediated AKT signaling pathway, we found that PVT1 knockdown significantly decreased the protein level of VEGFA and its downstream p-AKT, but not the p-ERK (extracellular signal-regulated kinase) or p-JNK (c-Jun N-terminal kinase) (Figures 6B and 6C). Furthermore, we found that agomir-16-5p could significantly decrease the expression of VEGFA, p-VEGFR1, p-AKT, and eNOS (endothelial nitric oxide synthase) (Figures 6D and 6E). Conversely, antagomir-16-5p upregulated the expression of these proteins (Figures 6D and 6E). These results are consistent with recent studies that report that miR-16-5p could directly bind to the 3′ UTR of VEGFA to activate the VEGFA/AKT signaling pathway.33,34
Next, the specific VEGFA silenced and overexpressed plasmids were used to further explore whether PVT1 knockdown suppressed CRC progression via VEGFA/VEGFR1 inactivation. The results showed that VEGFA silenced plasmids markedly repressed the phosphorylation of VEGFA, and then the downstream targeted genes VEGFR1, AKT, and eNOS were all inhibited (Figures 6F and 6G).
Finally, we treated PVT1 knockdown HCT116 and SW480 cells with agomir-16-5p and antagomir-16-5p separately. We found that PVT1 knockdown and agomir-16-5p treatment could most significantly inhibit the expression of VEGFA, p-VEGFR1, p-AKT, and eNOS compared with the control and NC group (Figures 6H and 6I). However, PVT1 knockdown and the antagomir-16-5p treatment group could partially restore the expression of VEGFA, p-VEGFR1, p-AKT, and eNOS (Figures 6H and 6I). Laser scanning confocal microscopy results further confirmed Western blot results (Figures S3 and S4). Taken together, these results reveal that the miR-16-5p/VEGFA/VEGFR1/AKT axis was essential for PVT1-mediated oncogenic functions.
Knockdown of PVT1 Combined with miR-16-5p Overexpression Suppressed Tumor Growth and Metastasis in Xenograft Animal Models
To further assess whether PVT1 exerts a tumor-promoting effect in vivo, we established a xenograft nude mouse model by subcutaneously injecting the shRNA-PVT1-transfected and/or agomir-16-treated HCT116 cells (n = 5 for each group). The untreated HCT116 cell group served as a control. As shown in Figures 7A, 7D, and 7E, sh-RNA PVT1 or agomir-16-5p markedly reduced tumor volume and weight, respectively. More importantly, the combined sh-RNA PVT1 and agomir-16-5p group displayed less tumor volume and weight than did that of sh-RNA PVT1 or agomir-16-5p alone.
Figure 7.
Knockdown of PVT1 Coupled with miR-16-5p Overexpression Suppressed Tumor Growth and Metastasis in Xenograft Animal Models
(A) Images of the xenograft model in BALB/c nude mice from four treatment groups (n = 5 for each group) on the 35th day. (B) Images of H&E staining and IHC staining for Ki-67, CD34, MMP-2, and MMP-9. Scale bar, 50 μm. (C) The levels of Ki-67, CD34, MMP-2, and MMP-9 were calculated. ∗∗∗p < 0.001 versus control; ###p < 0.001 versus shRNA-PVT1-2. (D) Tumor volumes were monitored with digital calipers during the time course of 5 weeks. ∗∗p < 0.01 versus control; ##p < 0.01 versus shRNA-PVT1-2. (E) Weights of tumors were measured at the end of this study. ∗∗p < 0.01 versus control; ##p < 0.01 versus shRNA-PVT1-2. T indicates error bars. Data are listed as mean ± SD of at least three independent experiments.
Immunohistochemistry (IHC) analysis also showed decreased Ki-67, CD34, MMP2, and MMP9 in the sh-RNA PVT1 + agomir-16-5p group compared with shRNA PVT1 or the agomir-16-5p group alone (Figures 7B and 7C). Overall, these data indicate that silencing of PVT1 suppresses CRC growth and metastasis in vivo, and silencing of PVT1 combined with miR-16-5p overexpression exhibits an additive inhibition of tumor growth.
Discussion
Extensive evidence has shown that lncRNAs can play a crucial role in tumorigenesis and growth development. lncRNA PVT1 was originally discovered as an activator of MYC in murine plasmacytoma variant translocations.35,36 Recently, accumulating studies have found that PVT1 played a crucial role in CRC, but the exact mechanism remains unclear.16,17 In the present study, PVT1 was found to be upregulated in CRC tissues and cell lines, and it was closely correlated with poor prognosis with bad clinicopathological features. Furthermore, cell growth and invading and migration ability were inhibited in CRC cells after PVT1 was knocked down. These data indicated that PVT1 functioned as an oncogene in CRC.
Increasing studies have demonstrate that lncRNAs regulate mRNA levels by competing for miRNAs, by acting as ceRNAs.23,37 Zhao et al.13 have reported that lncRNA PVT1 functions as sponge for miR-448 to promote pancreatic ductal adenocarcinoma development. Similarly, a recent study by Shen et al.38 revealed that lncRNA PVT1 could bind to miR-145 and then upregulate its target gene FSCN1 to promote esophageal carcinoma (EC) cell migration and invasion and inhibit apoptosis. Additionally, in CRC, He et al.16 found that PVT1 is upregulated in human CRC tissues, and in CRC cells, upregulated PVT1 stabilized the expression of Lin28 by harboring miR-128, which eventually repressed the Let7 family, the downstream target of Lin28. Chai et al.39 reported that PVT1 promoted CRC development by sponging and inhibiting miR-455 to elevate RUNX2 expression. In addition, an in vitro study by Shang et al.40 confirmed that lncRNA PVT1 may mediate the progression of CRC by regulating IRS1 via sponging miR-214-3p. In our study, bioinformatics analysis indicated that PVT1 could anneal with miR-16-5p, and a luciferase reporter assay and RNA pull-down assay were performed to further verify such interaction. We also found that PVT1 expression levels were markedly reversely correlated with the expression of miR-16-5p in CRC tissues and cells. Furthermore, PVT1 knockdown combined with miR-16-5p overexpression significantly suppressed the malignant characteristics of CRC cells. Downregulation of miR-16-5p could partially rescue the inhibition induced by PVT1 knockdown in both in vitro cell experiments and in vivo mouse subcutaneous xenograft models. Taken together, these data strongly supported the role of PVT1 as a miR-16-5p sponge in CRC; furthermore, the suppression of malignancy caused by PVT1 knockdown might result from the upregulation of miR-16-5p.
It is currently understood that miRNAs exert their function by modulating their target genes via elevating translational repression or mRNA degradation. miR-16-5p has been reported to be aberrantly expressed in various malignancies, including osteosarcoma,41 breast cancer,42 prostate cancer,43 and other tumor types. In our current study, we observed that miR-16-5p is downregulated in CRC cell lines and tissues. Overexpression of miR-16-5p inhibits the malignant behaviors of CRC cells, while the opposite effects are found in CRC cells with downregulated miR-16-5p, with these results being consistent with research results previously described.44
Angiogenesis, as a key cancer hallmark, is essential for tumorigenesis, progression, and prognosis. VEGFA, which is a key regulator of angiogenesis and migration, has been confirmed that it’s 3′ UTR has a specific direct binding site (nucleotides 195–217)33 of miR-16-5p in multiple myeloma and large-cell lymphoma.33,34 However, in colon cancer there remain few published data concerning whether miR-16-5p affects the expression of VEGFA. In the present study, we found that miR-16-5p overexpression reduced the expression of VEGFA on protein levels, and this result was opposite when downregulating miR-16-5p. The VEGF family of ligands includes VEGF-A, -B, -C, -D, and -E and placenta growth factor (PlGF). These ligands bind to three tyrosine kinase receptors: VEGFR1, VEGFR2, and VEGFR3. The extracellular ligand-binding domains exist on these three receptors, which could induce intracellular signaling upon binding and receptor dimerization. Of the three VEGFR tyrosine kinases, VEGFR2 is the best characterized to date. However, the studies performed by Fan et al.45 and Bhattacharya et al.32 have demonstrated that CRC cells, including HCT116 and SW480 cells, did not express VEGFR2. In the study by Bhattacharya et al.,32 they found that depletion of VEGFR1 in multiple CRC cell lines, including HCT116 and SW480 cells, led to strong inhibition of invasion and migration. Considering these findings, we propose that in CRC cells that lack VEGFR2, the VEGFA-VEGFR1 signaling pathway may regulate the angiogenesis and migration of CRC cells.
VEGFA and VEGFR1 together activate downstream signaling, including activation of the ERK1/2, JNK MAPK, and the phosphoinositide 3-kinase/AKT pathways.45 Previous studies have reported that AKT signaling pathway plays a critical role in various biological processes of cancers, such as proliferation, metastasis, survival, and angiogenesis.46, 47, 48 As is known, AKT is the downstream target of VEGFA; subsequently, we further examined whether PVT1 knockdown inhibits CRC tumor angiogenesis through the VEGFA/VEGFR1/AKT signaling pathway. Our data showed that PVT1 knockdown could significantly reduce the expression of VEGFA/VEGFR1 and the downstream AKT signaling pathway but neither the ERK1/2 nor JNK MAPK pathways. Therefore, we can speculate that PVT1 may stimulate the occurrence, progression, and metastasis of CRC through the miR-16-5p/VEGFA/VEGFR1/AKT signaling pathway.
To further verify our speculation, we subsequently proved that both agomir-16-5p and VEGFA small interfering RNA (siRNA) could restore the expression of VEGFA, VEGFR1, p-AKT, and eNOS. eNOS is one isoform of the NOS family, mainly found in endothelial cells, where it physically promotes vasodilation.49 Conversely, evidence also indicates that eNOS is expressed in cancer, where it plays a role in cell proliferation, antiapoptosis, angiogenesis, invasion, and metastasis.50, 51, 52, 53, 54 Furthermore, phosphorylation of eNOS and NO production can be upregulated by the AKT pathway.55 In the present study, we further found that agomir-16-5p could aggravate the VEGFA and VEGFR1 downregulation induced by PVT1 knockdown, and then attenuate the activation of AKT and its downstream target gene eNOS. However, antagomir-16-5p leads to opposite effects. Finally, an in vivo study demonstrated similar effects of PVT1 and miR-16-5p as found in vitro. PVT1 knockdown combined with miR-16-5p overexpression resulted in the smallest tumor volume and the longest survival, as well as inhibition of angiogenesis and EMT.
Conclusion
In summary, lncRNA PVT1 expression is found to be increased in CRC cells and tissues. In CRC patients, upregulated PVT1 positively correlates with poor prognosis and bad clinicopathological features. Furthermore, we verified that PVT1 knockdown inhibited EMT and the angiogenesis process, and it inactivated the miR-16-5p/VEGFA/VEGFR1/AKT signaling pathway in CRC (Figure S5). Our data will provide new insights into the underlying mechanism of CRC progression, and they suggest that PVT1 might serve as a potential prognostic biomarker and promising therapeutic target for CRC.
Materials and Methods
Patients and Clinical Samples
A total of 72 CRC patients (39 males and 33 females) between the ages of 43 and 72 years were enrolled in this study, each of whom underwent surgical treatments alone from August 2013 to September 2017 in Zhongda Hospital, Southeast University (Nanjing, China). None of these patients received chemotherapy or radiotherapy before or had other cancers besides CRC before. Sex and age have no special requirements in this study. CRC tissues and adjacent normal tissues of the 72 patients were collected. All samples were clinically and pathologically diagnosed. The use of human samples in this study were sanctioned by the local Ethics Committee at Zhongda Hospital, Southeast University (Nanjing, China, approval #2013ZDSYLL052-PO1), and in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Informed consent was obtained for experimentation with human subjects. The privacy rights of human subjects must always be observed. The patient characteristics are shown in Table 1. The identification of pathological characteristics was based on the American Joint Committee on Cancer (AJCC) staging criteria.56,57 Additionally, the carcinoembryonic antigen (CEA) results were obtained from the CRC patients’ blood tests performed before surgery at the hospital.
Cell Culture
Human normal colon epithelial cell (FHC), CRC cell lines (HCT116 and SW480), and human embryonic kidney cell line (HEK293T) were all obtained from American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM medium supplemented with 10% fetal bovine serum (Gibco, Vienna, Austria). Cells in this medium were placed in a humidified atmosphere of 5% CO2 at 37°C.
Quantitative Real-Time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, CA, USA) and was reverse transcribed to cDNAs via a reverse transcription kit (Takara, Dalian, China). Quantitative real-time RT-PCR was performed with SYBR Green (Takara, Dalian, China). GAPDH was used as reference for miRNAs or lncRNAs. The primers used in this study are given in Table S1. Quantitative real-time RT-PCR was conducted on an ABI 7500 system (Applied Biosystems, MA, USA). Thermal cycles were as follows: 95°C for 30 s, 95°C for 5 s for 40 cycles, and 60°C for 35 s.
Cell Transfection
Three sh-RNAs against the PVT1 gene were ligated into GV493 vectors, and the components hU6-MCS-CBh-gcGFP-IRES-puromycin (Genechem, Shanghai, China) and plasmid with a nontargeting sequence were used as a negative control (sh-NC). The three target sequences of sh-PVT1 were as follows: 5′-CCTGGGATTTAGGCACTTT-3′, 5′-GCTTCAACCCATTACGATT-3′, and 5′-CCTTCCAGTGGATTTCCTT-3′. Transfection was performed at nearly 60%–70% confluence of CRC cells using Polybrene reagents (Genechem, Shanghai, China). Stable cell lines were obtained after puromycin screening (Invitrogen, CA, USA). The efficiency of the transfected cell lines was tested using quantitative real-time RT-PCR.
Transient transfection of agomir-16-5p, antagomir-16-5p, agomir-NC, and antagomir-NC (GenePharma, Shanghai, China) were performed according to the manufacturer’s instructions for Lipofectamine 3000 (Life Technologies) reagents.
For the treatment, siRNAs against VEGFA and negative control siRNA were purchased from GenePharma (Shanghai, China). The VEGFA siRNA sequences were as follows: sense, 5′-GGCAGCUUGAGUUAAACGATT-3′; antisense, 5′-UCGUUUAACUCAAGCUGCCTT-3′.
To construct VEGFA overexpression plasmids, VEGFA cDNA was synthesized by GenePharma (Shanghai, China) and cloned into a pEX-1 (pG-CMV/MCS/EGFP/Neo) vector (GenePharma, Shanghai, China). The sequences were as follows: sense, 5′-ACCATGAACTTTCTGCTGTCTTGGGTGCAT-3′; antisense, 5′-TCACCGCCTCGGCTTGTCACATCTGCAAGT-3′. Transfection was carried out using Lipofectamine 3000 according to the manufacturer’s instructions.
Dual-Luciferase Reporter Assay
The PVT1 cDNA fragment containing the predicted potential miR-16-5p binding sites was amplified by PCR from human colon cancer tissues and then cloned into the GV272 luciferase reporter vector (GeneChem, Shanghai, China) to generate GV272-PVT1 (PVT1-WT). Also, the mutant miR-16-5p binding sites were replaced as indicated to generate GV272-PVT1-MUT (PVT1-MUT). T293 cells were co-transfected with 500 ng of PVT1-WT/MUT plasmid vector and 30 nM GV268-miR-16-5p vector plasmid or GV268-miR-16-5p NC vector plasmid by X-tremeGENE-HP (Roche, Basel, Switzerland). After 48 h of transfection, cells were harvested for luciferase reporter assay with a Dual-Luciferase assay kit (Promega).
Biotinylated RNA Pull-Down Assay
The pull-down assay with biotinylated RNA was performed as previously described.58,59 In brief, the biotinylated miR-16-5p probe was incubated with C-1 magnetic beads (Life Technologies, CA, USA) to generate probe-coated magnetic beads and then incubated with sonicated HCT116 and SW480 cells at 4°C overnight, followed by eluted and quantitative real-time RT-PCR. For miR-16-5p, pulled-down PVT1, HCT116, and SW480 cells with PVT1 overexpression were transfected with biotinylated miR-16-5p mimics or mutant using Lipofectamine 3000. The beads were washed five times with lysis buffer, and the bound RNAs in the pull-down materials were extracted using TRIzol reagent and analyzed by a quantitative real-time RT-PCR assay.
Cell Proliferation Assay
The proliferative ability of CRC cells was evaluated using a CCK-8 assay according to the manufacturer’s instruction (Dojindo Laboratories, Kumamoto, Japan). HCT116 and SW480 cells (1 × 103) were plated in 96-well plates and treated with 10 μL of CCK-8 solution and then spectrophotometrically analyzed by an automatic microplate reader at 450 nm (Synergy4; BioTek, Winooski, VT, USA).
Cell Cycle Analysis by Propidium Iodide Staining
For the cell cycle analysis, the transfected cells were collected and fixed in 70% ethanol overnight at −20°C and stained with propidium iodide (Kaiji, Nanjing, China) in a phosphate-buffered saline (PBS) solution containing R-Nase following the manufacturer’s instructions. The data were analyzed using ModFit 3.3 software (BD Biosciences, Sparks, MD, USA).
Cell Colony Formation Assay
For the colony formation assay, stable transfected cells were screened with puromycin, counted using a cell counting plate, and then seeded into six-well plates (Corning) with a density of 500 cells per well. Then, cells were cultured for 2 weeks. The cell colonies were then fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 10 min. The stained colonies were counted and imaged.
Scratch Wound Healing Assay
Cells were seeded at 4 × 104 cells/cm2. After 48 h, a straight scratch was made with a 200-μL pipette tip and the wound was imaged under the microscope. After 48 h, cells were imaged under the microscope and the area of the remaining scratch was calculated.
Hematoxylin and Eosin (H&E) and IHC Staining
Paraffin-block sections of subcutaneous xenografts in mice were stained with H&E and subsequently evaluated blindly by a pathologist. In addition, immunohistochemistry was performed using antibodies for Ki-67 (27309-1-AP, Proteintech, USA), CD34 (14486-1-AP, Proteintech, USA), MMP9 (10375-2-AP, Proteintech, USA), and MMP2 (10373-2-AP, Proteintech, USA) according to the manufacturer’s instructions. All sections were scored by the semiquantitative H score approach and validated by two experienced pathologists.
Laser Scanning Confocal Microscopy
The transfected cells were harvested and washed with PBS and then fixed in fresh paraformaldehyde (4%) for 20 min at 37°C. Then, they were treated with Triton X-100 (0.5%) to increase permeability, followed by washing with PBS. After that, the cells were incubated with rabbit or mouse anti-human eNOS (18985-1-AP, Proteintech, USA), p-AKT (66444-1-lg, Proteintech, USA), VEGFA (ab52917, Abcam, Cambridge, MA, USA), or p-VEGFR1 (AF4170, R&D Systems, USA) antibody for 3 h at room temperature. The cells were washed using PBS followed by incubation with goat anti-rabbit/anti-mouse Cy3 or fluorescein isothiocyanate (FITC) antibody for 60 min at room temperature in dark. The cells were then subjected to confocal microscopy for capturing images.
Western Blotting
The HCT116 and SW480 cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer. Then, equal amounts of protein were resolved by SDS-PAGE analysis and electrotransferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Schwalbach, Germany), then blocked with 5% skim milk powder and incubated with primary antibody at 4°C overnight. The primary antibodies used were anti-VEGFA (ab52917, Abcam, Cambridge, MA, USA), anti-VEGFR1 (AF4170, R&D Systems, USA), anti-p-VEGFR1 (Try1213, Millipore, USA), anti-AKT (10176-2-AP, Proteintech, USA), anti-p-AKT (66444-1-lg, Proteintech, USA), and anti-eNOS (18985-1-AP, Proteintech, USA). Then, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature and the blots were visualized using an enhanced chemiluminescence kit (Pierce, Waltham, MA, USA).
Animal Experiments
For the xenograft tumor model, 5-week-old male BALB/c nude mice, which were purchased from the Yangzhou University Animal Center, were randomly divided into four groups (n = 5 for each group). HCT116 control and post-transfected cells (5 × 106/0.2 mL of PBS cells) were subcutaneously inoculated into the thigh root of the right hindlimb of each nude mouse. Tumor volumes were measured every 4 days with digital calipers and were calculated by the following formula: tumor volume = ½(length × width2). 35 days later, under general anesthesia, tumors were excised, weighed, processed for paraffin embedding, and stored in liquid nitrogen. Then, the tumor tissues were fixed in 10% neutral phosphate-buffered formalin, followed by H&E and IHC staining. Then, all of the lungs and livers were dissected and separated to observe whether distant metastasis was present. The animal experiment protocols were completed in compliance with the Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Welfare Committee (acceptance #20181130007).
Statistical Analysis
All statistical analyses were carried out using SPSS 19.0 statistical software (IBM, Chicago, IL, USA), and figures were produced using GraphPad Prism 7.0. All data are presented as the mean ± SD of at least three independent experiments. Differences between the groups were tested using a Student’s t test or one-way ANOVA. A chi-square test was performed to compare the correlation between PVT1 level and clinical parameters. Survival curves were plotted by the Kaplan-Meier method and compared statistically using the log rank test. Risk score analysis was performed to evaluate the effectiveness of PVT1 for prediction. The correlations were analyzed using Pearson’s correlation analysis. The differences were considered to be significant at p < 0.05.
Author Contributions
G.W. and W.Z. designed the study. R.S. provided suggestions for the project. H.W. and M.W. performed the primary experiments and wrote the manuscript. X.J., W.X., and X.F. analyzed the data. R.Z., C.D., and F.Z. revised the figures and tables. X.S., Z.Z., and J.T. collected the tissue samples and clinical data of CRC patients. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We acknowledge Guoqing Wang, PhD and FenGan Ding, PhD (Department of Pathology, Zhongda Hospital, Southeast University, Nanjing, China) for excellent technical assistance. This work was supported by grants from the Postgraduate Research and Practice Innovation Program of Jiangsu Province (no. KYCX18_0178); the National Natural Science Foundation of China (no. 81773624, 81600415, 81600429, 81603016, and 81570503); the National Science and Technology Major Project (nos. 2018ZX09301026-005 and 2020ZX09201015); and the National Natural Science Foundation of Jiangsu Province (nos. BE2017746 and BK20160706).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.03.006.
Contributor Information
Weijia Zhang, Email: weijia.zhang@mssm.edu.
Guoqiu Wu, Email: nationball@163.com.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Dienstmann R., Vermeulen L., Guinney J., Kopetz S., Tejpar S., Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 2017;17:268. doi: 10.1038/nrc.2017.24. [DOI] [PubMed] [Google Scholar]
- 3.Aguilo F., Zhou M.M., Walsh M.J. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Z., Zhu Z., Zhang B., Li W., Li X., Wu X., Wang L., Fu L., Fu L., Dong J.T. Frequent mutation of rs13281615 and its association with PVT1 expression and cell proliferation in breast cancer. J. Genet. Genomics. 2014;41:187–195. doi: 10.1016/j.jgg.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y.R., Zang S.Z., Zhong C.L., Li Y.X., Zhao S.S., Feng X.J. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 6.Ding C., Yang Z., Lv Z., Du C., Xiao H., Peng C., Cheng S., Xie H., Zhou L., Wu J., Zheng S. Long non-coding RNA PVT1 is associated with tumor progression and predicts recurrence in hepatocellular carcinoma patients. Oncol. Lett. 2015;9:955–963. doi: 10.3892/ol.2014.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding J., Li D., Gong M., Wang J., Huang X., Wu T., Wang C. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. OncoTargets Ther. 2014;7:1625–1630. doi: 10.2147/OTT.S68854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi Y., Sawada G., Kurashige J., Uchi R., Matsumura T., Ueo H., Takano Y., Eguchi H., Sudo T., Sugimachi K. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br. J. Cancer. 2014;110:164–171. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D., Li Y., Zhang H., Hu X. Knockdown of Lncrna PVT1 enhances radiosensitivity in non-small cell lung cancer by sponging Mir-195. Cell. Physiol. Biochem. 2017;42:2453–2466. doi: 10.1159/000480209. [DOI] [PubMed] [Google Scholar]
- 10.Kong R., Zhang E.B., Yin D.D., You L.H., Xu T.P., Chen W.M., Xia R., Wan L., Sun M., Wang Z.X. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou D.D., Liu X.F., Lu C.W., Pant O.P., Liu X.D. Long non-coding RNA PVT1: emerging biomarker in digestive system cancer. Cell Proliferat. 2017;50:e12398. doi: 10.1111/cpr.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshida K., Toden S., Ravindranathan P., Han H., Goel A. Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis. 2017;38:1036–1046. doi: 10.1093/carcin/bgx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao L., Kong H., Sun H., Chen Z., Chen B., Zhou M. lncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J. Cell. Physiol. 2018;233:4044–4055. doi: 10.1002/jcp.26072. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y., Du H., Bao L., Liu W. lncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol. Med. 2018;15:238–250. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J., Li Y., Sang Y., Yu B., Lv D., Zhang W., Feng H. lncRNA PVT1 regulates triple-negative breast cancer through KLF5/beta-catenin signaling. Oncogene. 2018;37:4723–4734. doi: 10.1038/s41388-018-0310-4. [DOI] [PubMed] [Google Scholar]
- 16.He F., Song Z., Chen H., Chen Z., Yang P., Li W., Yang Z., Zhang T., Wang F., Wei J. Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene. 2019;38:164–179. doi: 10.1038/s41388-018-0432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng Y.Y., Moriarity B.S., Gong W., Akiyama R., Tiwari A., Kawakami H., Ronning P., Reuland B., Guenther K., Beadnell T.C. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson D.W., Dinger M.E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 19.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 21.Lin X., Wang S., Sun M., Zhang C., Wei C., Yang C., Dou R., Liu Q., Xiong B. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019;12:20. doi: 10.1186/s13045-019-0708-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Ullmann P., Nurmik M., Schmitz M., Rodriguez F., Weiler J., Qureshi-Baig K., Felten P., Nazarov P.V., Nicot N., Zuegel N. Tumor suppressor miR-215 counteracts hypoxia-induced colon cancer stem cell activity. Cancer Lett. 2019;450:32–41. doi: 10.1016/j.canlet.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 23.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D., Li Y., Luo G., Xiao X., Tao D., Wu X., Wang M., Huang C., Wang L., Zeng F., Jiang G. lncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Peng W., Si S., Zhang Q., Li C., Zhao F., Wang F., Yu J., Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J. Exp. Clin. Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Z., Zhang A., Liu S., Lu F., Guo Y., Zhang G., Xu F., Shi Y., Shen S., Liang J., Guo W. Aberrant methylation-mediated silencing of lncRNA MEG3 functions as a ceRNA in esophageal cancer. Mol. Cancer Res. 2017;15:800–810. doi: 10.1158/1541-7786.MCR-16-0385. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y., Shen L., Li F., Yang J., Wan X., Ouyang M. MicroRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J. Cell. Physiol. 2019;234:21380–21394. doi: 10.1002/jcp.28747. [DOI] [PubMed] [Google Scholar]
- 28.Ostenfeld M.S., Jensen S.G., Jeppesen D.K., Christensen L.L., Thorsen S.B., Stenvang J., Hvam M.L., Thomsen A., Mouritzen P., Rasmussen M.H. miRNA profiling of circulating EpCAM+ extracellular vesicles: promising biomarkers of colorectal cancer. J. Extracell. Vesicles. 2016;5:31488. doi: 10.3402/jev.v5.31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madadi S., Soleimani M. Comparison of miR-16 and cel-miR-39 as reference controls for serum miRNA normalization in colorectal cancer. J. Cell. Biochem. 2019;120:4802–4803. doi: 10.1002/jcb.28174. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z., Zhao C., Wang L., Cao X., Li J., Huang R., Lao Q., Yu H., Li Y., Du H. A VEGFR1 antagonistic peptide inhibits tumor growth and metastasis through VEGFR1-PI3K-AKT signaling pathway inhibition. Am. J. Cancer Res. 2015;5:3149–3161. [PMC free article] [PubMed] [Google Scholar]
- 31.Mezquita B., Pineda E., Mezquita J., Mezquita P., Pau M., Codony-Servat J., Martínez-Balibrea E., Mora C., Maurel J., Mezquita C. LoVo colon cancer cells resistant to oxaliplatin overexpress c-MET and VEGFR-1 and respond to VEGF with dephosphorylation of c-MET. Mol. Carcinog. 2016;55:411–419. doi: 10.1002/mc.22289. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharya R., Fan F., Wang R., Ye X., Xia L., Boulbes D., Ellis L.M. Intracrine VEGF signalling mediates colorectal cancer cell migration and invasion. Br. J. Cancer. 2017;117:848–855. doi: 10.1038/bjc.2017.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun C.Y., She X.M., Qin Y., Chu Z.B., Chen L., Ai L.S., Zhang L., Hu Y. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 2013;34:426–435. doi: 10.1093/carcin/bgs333. [DOI] [PubMed] [Google Scholar]
- 34.Dejean E., Renalier M.H., Foisseau M., Agirre X., Joseph N., de Paiva G.R., Al Saati T., Soulier J., Desjobert C., Lamant L. Hypoxia-microRNA-16 downregulation induces VEGF expression in anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphomas. Leukemia. 2011;25:1882–1890. doi: 10.1038/leu.2011.168. [DOI] [PubMed] [Google Scholar]
- 35.Webb E., Adams J.M., Cory S. Variant (6;15) translocation in a murine plasmacytoma occurs near an immunoglobulin kappa gene but far from the myc oncogene. Nature. 1984;312:777–779. doi: 10.1038/312777a0. [DOI] [PubMed] [Google Scholar]
- 36.Cory S., Graham M., Webb E., Corcoran L., Adams J.M. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 1985;4:675–681. doi: 10.1002/j.1460-2075.1985.tb03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen S.N., Li K., Liu Y., Yang C.L., He C.Y., Wang H.R. Down-regulation of long noncoding RNA PVT1 inhibits esophageal carcinoma cell migration and invasion and promotes cell apoptosis via microRNA-145-mediated inhibition of FSCN1. Mol. Oncol. 2019;13:2554–2573. doi: 10.1002/1878-0261.12555. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Chai J., Guo D., Ma W., Han D., Dong W., Guo H., Zhang Y. A feedback loop consisting of RUNX2/LncRNA-PVT1/miR-455 is involved in the progression of colorectal cancer. Am. J. Cancer Res. 2018;8:538–550. [PMC free article] [PubMed] [Google Scholar]
- 40.Shang A.Q., Wang W.W., Yang Y.B., Gu C.Z., Ji P., Chen C., Zeng B.J., Wu J.L., Lu W.Y., Sun Z.J., Li D. Knockdown of long noncoding RNA PVT1 suppresses cell proliferation and invasion of colorectal cancer via upregulation of microRNA-214-3p. Am. J. Physiol. Gastrointest. Liver Physiol. 2019;317:G222–G232. doi: 10.1152/ajpgi.00357.2018. [DOI] [PubMed] [Google Scholar]
- 41.Maximov V.V., Akkawi R., Khawaled S., Salah Z., Jaber L., Barhoum A., Or O., Galasso M., Kurek K.C., Yavin E., Aqeilan R.I. miR-16-1-3p and miR-16-2-3p possess strong tumor suppressive and antimetastatic properties in osteosarcoma. Int. J. Cancer. 2019;145:3052–3063. doi: 10.1002/ijc.32368. [DOI] [PubMed] [Google Scholar]
- 42.Youness R.A., Hafez H.M., Khallaf E., Assal R.A., Abdel Motaal A., Gad M.Z. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell. Physiol. 2019;234:20286–20297. doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 43.Wang F., Mao A., Tang J., Zhang Q., Yan J., Wang Y., Di C., Gan L., Sun C., Zhang H. MicroRNA-16-5p enhances radiosensitivity through modulating cyclin D1/E1-pRb-E2F1 pathway in prostate cancer cells. J. Cell. Physiol. 2019;234:13182–13190. doi: 10.1002/jcp.27989. [DOI] [PubMed] [Google Scholar]
- 44.Liu R., Lu Z., Gu J., Liu J., Huang E., Liu X., Wang L., Yang J., Deng Y., Qian J. MicroRNAs 15A and 16-1 activate signaling pathways that mediate chemotaxis of immune regulatory B cells to colorectal tumors. Gastroenterology. 2018;154:637–651.e7. doi: 10.1053/j.gastro.2017.09.045. [DOI] [PubMed] [Google Scholar]
- 45.Fan F., Wey J.S., McCarty M.F., Belcheva A., Liu W., Bauer T.W., Somcio R.J., Wu Y., Hooper A., Hicklin D.J., Ellis L.M. Expression and function of vascular endothelial growth factor receptor-1 on human colorectal cancer cells. Oncogene. 2005;24:2647–2653. doi: 10.1038/sj.onc.1208246. [DOI] [PubMed] [Google Scholar]
- 46.Mashreghi M., Azarpara H., Bazaz M.R., Jafari A., Masoudifar A., Mirzaei H., Jaafari M.R. Angiogenesis biomarkers and their targeting ligands as potential targets for tumor angiogenesis. J. Cell. Physiol. 2018;233:2949–2965. doi: 10.1002/jcp.26049. [DOI] [PubMed] [Google Scholar]
- 47.Wu W., Gao H., Li X., Zhu Y., Peng S., Yu J., Zhan G., Wang J., Liu N., Guo X. lncRNA TPT1-AS1 promotes tumorigenesis and metastasis in epithelial ovarian cancer by inducing TPT1 expression. Cancer Sci. 2019;110:1587–1598. doi: 10.1111/cas.14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu W., Yang B., Leng X., Pei X., Xu J., Liu M., Dong Q., Tao D., Lu Y., Liu Y., Yang Y. Testis-specific protein, Y-linked 1 activates PI3K/AKT and RAS signaling pathways through suppressing IGFBP3 expression during tumor progression. Cancer Sci. 2019;110:1573–1586. doi: 10.1111/cas.13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Wallerath T., Förstermann U. Physiological mechanisms regulating the expression of endothelial-type NO synthase. Nitric Oxide. 2002;7:132–147. doi: 10.1016/s1089-8603(02)00127-1. [DOI] [PubMed] [Google Scholar]
- 50.Lampson B.L., Kendall S.D., Ancrile B.B., Morrison M.M., Shealy M.J., Barrientos K.S., Crowe M.S., Kashatus D.F., White R.R., Gurley S.B. Targeting eNOS in pancreatic cancer. Cancer Res. 2012;72:4472–4482. doi: 10.1158/0008-5472.CAN-12-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punathil T., Tollefsbol T.O., Katiyar S.K. EGCG inhibits mammary cancer cell migration through inhibition of nitric oxide synthase and guanylate cyclase. Biochem. Biophys. Res. Commun. 2008;375:162–167. doi: 10.1016/j.bbrc.2008.07.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong X., Li H. eNOS protects prostate cancer cells from TRAIL-induced apoptosis. Cancer Lett. 2004;210:63–71. doi: 10.1016/j.canlet.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y., Yan W., Lu X., Qian C., Zhang J., Li P., Shi L., Zhao P., Fu Z., Pu P. Overexpression of osteopontin induces angiogenesis of endothelial progenitor cells via the avβ3/PI3K/AKT/eNOS/NO signaling pathway in glioma cells. Eur. J. Cell Biol. 2011;90:642–648. doi: 10.1016/j.ejcb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Ying L., Hofseth L.J. An emerging role for endothelial nitric oxide synthase in chronic inflammation and cancer. Cancer Res. 2007;67:1407–1410. doi: 10.1158/0008-5472.CAN-06-2149. [DOI] [PubMed] [Google Scholar]
- 55.Tanimoto T., Jin Z.G., Berk B.C. Transactivation of vascular endothelial growth factor (VEGF) receptor Flk-1/KDR is involved in sphingosine 1-phosphate-stimulated phosphorylation of Akt and endothelial nitric-oxide synthase (eNOS) J. Biol. Chem. 2002;277:42997–43001. doi: 10.1074/jbc.M204764200. [DOI] [PubMed] [Google Scholar]
- 56.Egner J.R. AJCC Cancer Staging Manual. JAMA. 2010;304:1726–1727. [Google Scholar]
- 57.Weiser M.R. AJCC 8th Edition: Colorectal Cancer. Ann. Surg. Oncol. 2018;25:1454–1455. doi: 10.1245/s10434-018-6462-1. [DOI] [PubMed] [Google Scholar]
- 58.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 59.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., Sun H., Pan Y., He B., Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.