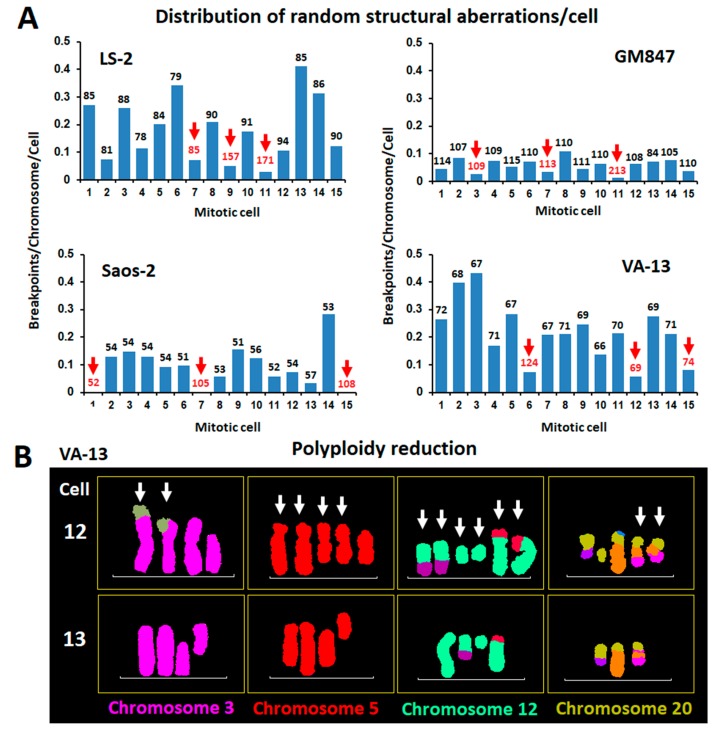
Figure 2.
Distribution of random structural chromosomal instability in neoplasia (CIN) among co-dividing alternative lengthening of telomeres (ALT) cells and the possible role of WGD in the perpetuation of the integrity of driver chromosomal rearrangements. (A) Uneven distribution of non-clonal structural chromosomal rearrangements between 15 randomly picked, co-dividing cells from 4 human ALT cell lines. Bars indicate structural CIN load, calculated as breakpoints/chromosome. Chromosome number/metaphase is indicated above each data bar. From every cell line we selected 3 cells with the lower rates of structural chromosomal instability. Interestingly, half of the 12 CIN escapee cells (red arrows and numbers) were byproducts of WGD or showed evidence of WGD reduction. (B) Pseudo-colored partial karyograms of two co-dividing VA-13 mitotic nuclei belonging to the major clones and each bearing 69 chromosomes. In contrast to #13, cell #12 displays extremely low rates of structural CIN and presents several duplicated copies of clonal recombinant chromosomes, suggesting WGD followed by multiple chromosome losses.