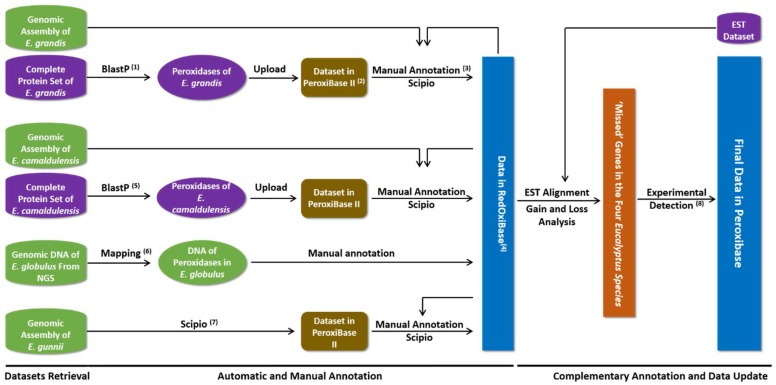
Figure 1.
Workflow of the comprehensive annotation process of Eucalyptus ROS genes in four Eucalyptus species. The comprehensive annotation, starting from the acquisition of data to the storage in a database, consists of automatic annotation, manual annotation and experimental detection as a complimentary annotation. The experimental detection for the missed sequences would only be possible when the relationship between target organisms is very close. (1) The query data for this BlastP is the protein sequence set of P. trichocarpa. (2) The ReroxiBase II is an assistant database of RedOxiBase, which is used for keeping some private temporary data during the annotation process. (3) The Scipio program takes the manually annotated protein sequences as the query. (4) The data stored in RedOxiBase contain protein, DNA, CDS sequences and the gene structure information for most of the records. Chromosomal positions are also included for the E. grandis genes. (5) The query for the BlastP similarity search is the protein set of E. grandis. (6) Mapping program takes the DNA set of E. grandis and E. camaldulensis as the query to obtain the peroxidase DNA sequences of E. globulus. (7) The protein set containing peroxidases of E. grandis, E. camaldulensis and E. globulus is used as the query for the Scipio program to obtain the peroxidase data. (8) PCR was performed for the “missing” genes among the organisms.