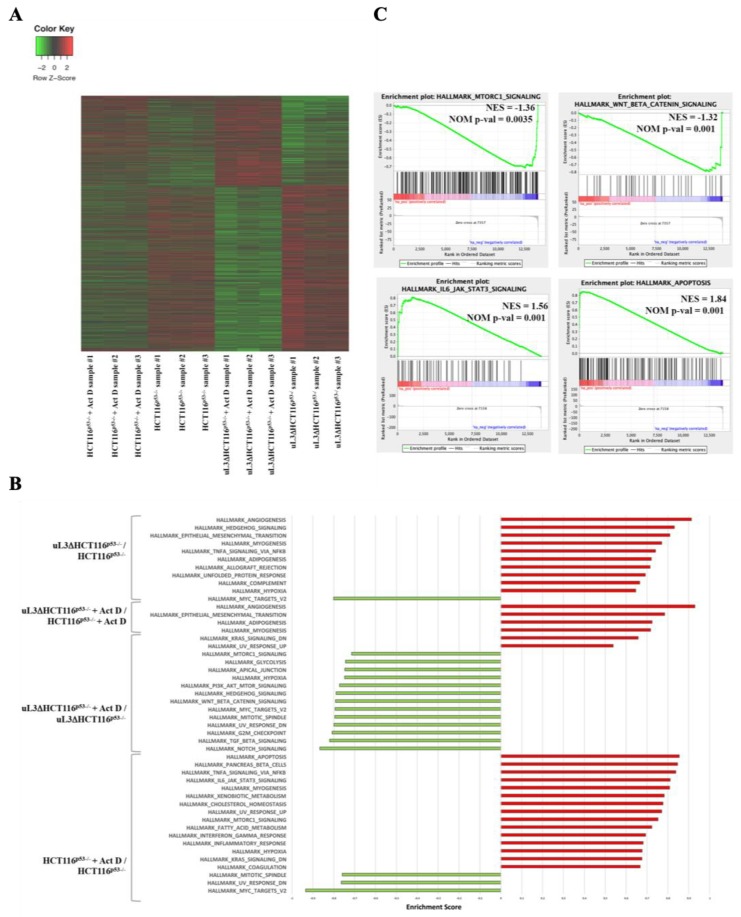
Figure 3.
Key genes and pathways regulated by Act D treatment in HCT 116p53−/− cells and uL3ΔHCT 116p53−/− cells. (A) Unsupervised hierarchical Z-Score clustering. Clustering analyses revealed differentially expressed genes (DEGs) between the experimental groups. The color scale means the gene expression standard deviations from the mean, with green for low expression and red for the high expression levels. (B) GSEA querying hallmark genes depicting significant enrichment of signaling pathway genes as cell metabolism, apoptosis, autophagy, cell growth, as well as cancer-related pathways. (C) Enrichment plots for gene sets related to IL6/JAK/STAT3 and apoptosis pathways significantly enriched in Act D-treated HCT116p53−/− cells versus untreated HCT116p53−/− cells and for the most negatively enriched pathways mTORC 1 and WNT_Beta_Catenin in uL3ΔHCT 116p53−/− cells after Act D treatment compared to deleted untreated cells. The top portion of each panel shows the normalized enrichment scores (NES) for each gene; the bottom portion of the plot indicates the value of the ranking metric moving down the list of ranked genes. Location of the gene set members are indicated by black lines in the center of the plot. The gray plot at the bottom represents the ranked list of all differentially expressed genes. Genes which are more highly expressed are in red whereas genes more highly expressed in blue. GSEA, Gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate.