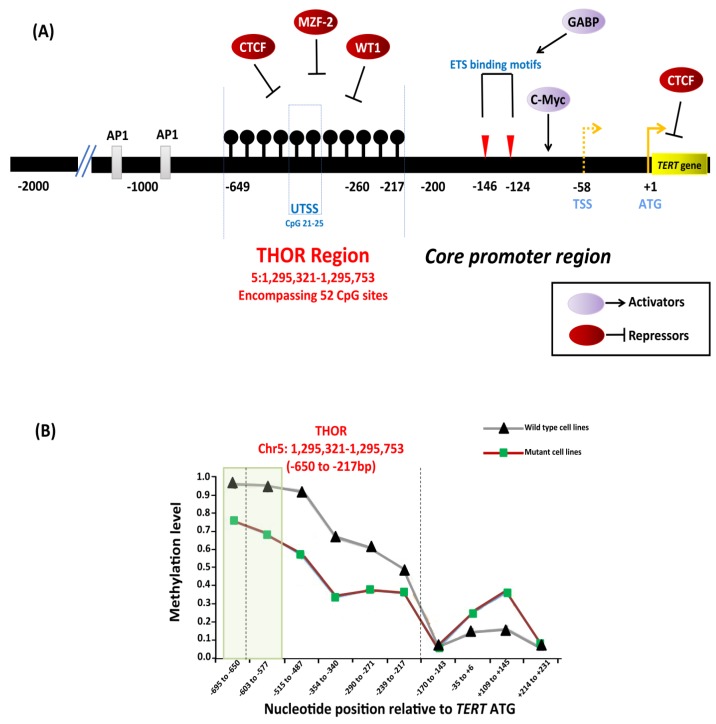
Figure 7.
Epigenetic regulation of TERT in cancers. (A) Depiction of transcription factors along with binding sites, TERT promoter mutations −124C > T and −146C > T, hypermethylated region upstream of transcription start site (THOR). Binding of transcriptional activators, c-Myc, and repressors, CCCTC-binding factor (CTCF), myeloid zinc finger protein-2 (MZF-2), and Wilms tumor 1 (WT1) to the TERT promoter is controlled by DNA methylation as methylated CpGs prevent the binding to the target sites leading to TERT activation. The black lollipops represent methylated CpG sites. (B) Relative DNA methylation in tumor-derived cell lines with and without TERT promoter mutations. The green box represents a specific region in THOR (−668 to −577 bp relative to ATG) that is shown to be less methylated in cell lines with TERT promoter mutations than in cell lines without mutations. Adapted from [123,146,147].