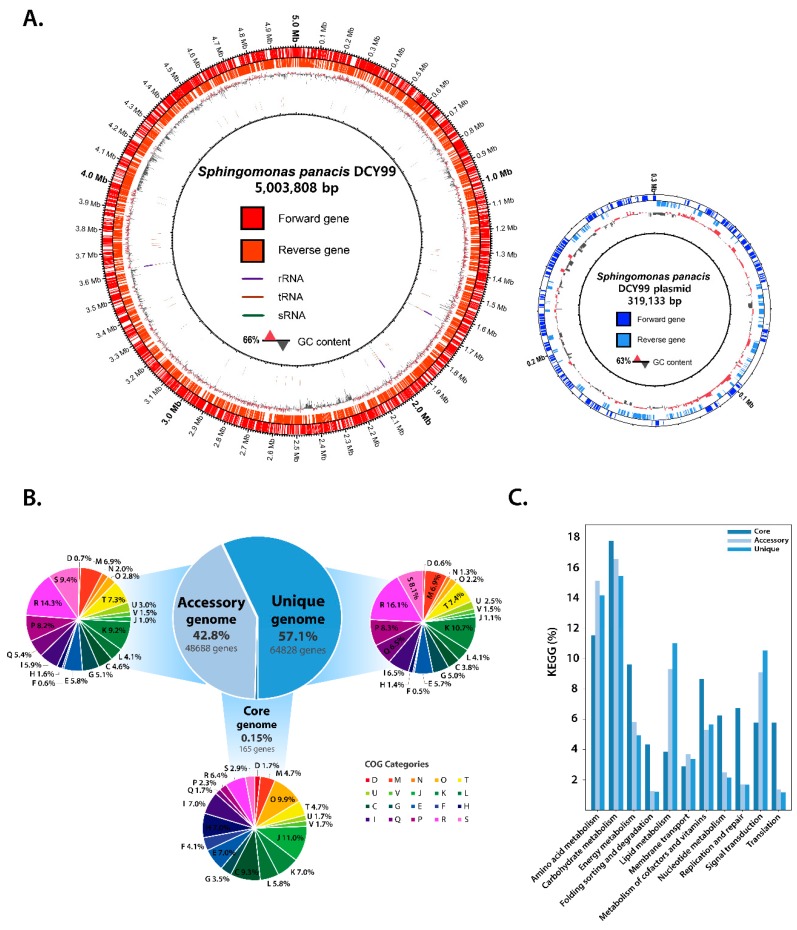
Figure 1.
(A) Visualization of the circular genome of S. panacis DCY99T. The genome is shown with a base pair (bp) ruler on the outer ring. The S. panacis DCY99T main chromosome is 5,003,808 bp in size; the plasmid is 319,133 bp in size. The chromosome is arranged clockwise. The two outer circles represent S. panacis DCY99T CDSs on the forward and reverse strands, respectively. The next circle indicates GC% content and next three circles indicate rRNA, tRNA, and sRNA, respectively. The S. panacis DCY99T plasmid does not contain the three RNAs. (B) Sphingomonas pan-genome statistics. The Sphingomonas pan-genome can be subdivided into three categories: (i) the core-genome (the set of genes shared by all genomes), (ii) the accessory-genome (the set of genes present in some but not all genomes), and (iii) the unique-genome (genes that are unique to a single genome). The function of each gene in a group was classified using COGs. COG categories are as follows. For cellular processes and signaling, D: cell cycle control, cell division, and chromosome partitioning; M: cell wall/membrane/envelope biogenesis, N: cell motility; O: posttranslational modification, chaperones and protein turnover, T: signal transduction mechanisms, U: intracellular trafficking, secretion, and vesicular transport, V: defense mechanisms. For information storage and processing, J: translation, ribosomal structure, and biogenesis, K: transcription, L: replication, recombination, and repair. For metabolism, C: energy production and conversion, G: carbohydrate transport and metabolism, E: amino acid transport and metabolism, F: nucleotide transport and metabolism, H: coenzyme transport and metabolism, I: lipid transport and metabolism, P: inorganic ion transport and metabolism, Q: secondary metabolites biosynthesis, transport, and catabolism, R: general function prediction only, and S: function unknown. (C) KEGG pathway distribution of the 159 Sphingomonas strains.