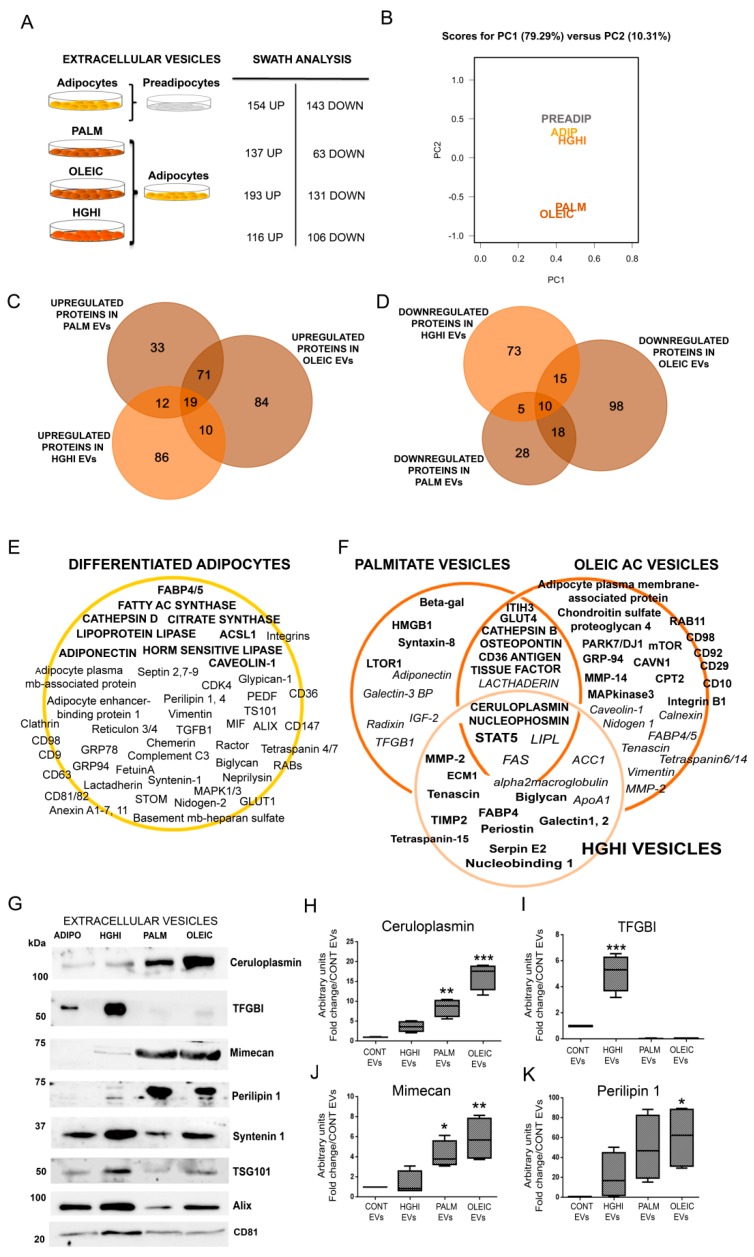
Figure 3.
Quantitative Sequential Window Acquisition of All Theoretical Mass Spectra (SWATH) analysis of EVs shed by lipid hypertrophied and insulin resistant adipocytes identifies obesity-related proteins and potential biomarkers. Differences on protein cargo of adipocytes vs. preadipocytes, and also in those vesicles shed by palmitate, oleic, and HGHI treated cells compared to control adipocytes by comparative analysis of EVs by Sequential Window Acquisition of all theoretical fragment-ion spectra is indicated (A). Principal component analysis (PCA) analysis of transformed SWATH areas for the quantitative comparison of all samples (n = 4 independent experiments; n = 3 technical replicates for each independent experiment) (B). Venn diagrams comparing those proteins upregulated (C) and downregulated (D) in EVs from the three pathologic cell models is shown. A representative proteome map of EVs secreted from control adipocytes compared to non-differentiated cells is represented showing representative upregulated proteins in bold (E). A proteome map highlighting selected proteins up (bold) and down (italic) regulated in EVs isolated from lipid hypertrophied (palmitate/oleic acid) and insulin resistant (HGHI) adipocytes (F). Representative images of immunoblots and band quantification in box plot graphs for cereluplasmin, transforming growth factor-beta-induced protein ig-h3 (TFGBI), mimecan, perilipin 1, syntenin 1, TSG101, Alix, and CD81 in independent isolated vesicles from all the cell models is shown (n = 4 independent lysates) (G–K); differences were assayed by one-way Anova-Kruskall Wallis test followed by Dunn’s multiple comparison test (p ≤ 0.05 was considered statistically significant: * p < 0.05, ** p < 0.01, and *** p < 0.001).