Abstract
In colorectal cancer (CRC), upregulation of the C-X-C motif chemokine receptor 4 (CXCR4) is correlated with metastasis and poor prognosis, highlighting the need to further elucidate CXCR4’s regulation in CRC. For the first time, DNA methylation and 5-hydroxymethylcytosine aberrations were investigated to better understand the epigenetic regulation of CXCR4 in CRC. CXCR4 expression levels were measured using qPCR and immunoblotting in normal colon tissues, primary colon cancer tissues and CRC cell lines. Publicly available RNA-seq and methylation data from The Cancer Genome Atlas (TCGA) were extracted from tumors from CRC patients. The DNA methylation status spanning CXCR4 gene was evaluated using combined bisulfite restriction analysis (COBRA). The methylation status in the CXCR4 gene body was analyzed using previously performed nano-hmC-seal data from colon cancers and adjacent normal colonic mucosa. CXCR4 expression levels were significantly increased in tumor stromal cells and in tumor colonocytes, compared to matched cell types from adjacent normal-appearing mucosa. CXCR4 promoter methylation was detected in a minority of colorectal tumors in the TCGA. The CpG island of the CXCR4 promoter showed increased methylation in three of four CRC cell lines. CXCR4 protein expression differences were also notable between microsatellite stable (MSS) and microsatellite instable (MSI) tumor cell lines. While differential methylation was not detected in CXCR4, enrichment of 5-hydroxymethylcytosine (5hmC) in CXCR4 gene bodies in CRC was observed compared to adjacent mucosa.
Keywords: colorectal cancer, DNA methylation, epigenetic regulation, CXCR4 gene expression, 5-hydroxymethylcytosine
1. Introduction
C-X-C motif chemokine ligand 12 (CXCL12), and its cognate receptor C-X-C motif chemokine receptor 4 (CXCR4), constitutes a major cytokine-signaling pathway involved in both health and disease [1,2]. CXCR4 deletion is embryonic lethal, emphasizing its essential role in normal development [3]. In addition, CXCR4 overexpression promotes tumorigenesis and its expression and activity are associated with cell proliferation [4,5,6,7,8], invasion [8,9,10,11,12,13], migration [8,14,15,16,17,18,19,20], inflammation [21], angiogenesis [22], and metastasis [23,24,25,26,27,28,29,30] in several cancer models. These myriad roles reflect the fact that CXCR4/CXCL12 signaling pathways can be activated by both autocrine and paracrine signaling pathways (e.g., via stromal (CXCL12)-epithelial (CXCR4) crosstalk). In the case of tumor metastasis, CXCL12 is released from stromal cells within the metastatic microenvironment, which in turn activates CXCR4 on cancer cells to increase cancer cell adhesion, motility and invasion in colon cancer cells [8]. Indeed, CXCR4 expression is an independent prognostic marker for several malignancies including: AML, multiple myeloma, squamous cell carcinoma, gastric cancer, renal cancer, hepatocellular carcinoma, and colorectal cancer (CRC) [31,32,33,34,35,36,37]. In the case of CRC patients, a meta-analysis correlated high CXCR4 expression with liver and lymph node metastasis and ultimately a reduction in overall survival [38].
Although the functions of CXCR4 are well described in tumor growth and invasion, the regulation of CXCR4 gene expression in cancer is less well understood. While upstream loss-of-function mutations of the von Hippel-Lindau tumor suppressor gene (VHL) or aberrant signaling may result in CXCR4 overexpression in some cancers [39,40], other studies point to epigenetic regulation of CXCR4. For example, cytosine methylation (5mC) occurs widely in cancers including CRC [41,42,43,44,45,46]. Prior studies have linked increased CXCR4 promoter cytosine methylation (5mC) to decreased CXCR4 protein expression in several non-colonic cancers such as primary breast cancer [29], decreased mRNA and protein in melanoma cell lines [47] and primary cervical cancer [48]. Increased CXCR4 expression was achieved with the knock-down of DNA methyltransferase 1 (DNMT1) or DNA methyltransferase 3 beta (DNMT3B), or with the inhibition of DNA methylation by 5-aza-2-deoxycytidine (5-aza) in pancreatic cancer cells [49]. Similarly, 5-aza treatment increased CXCR4 mRNA and protein levels in melanoma cells, and concomitantly enhanced cell migration [47]. These studies suggest that 5mC is an epigenetic silencer of CXCR4 in cancer. Another epigenetic marker is 5-hydroxymethylcytosine (5hmC), which is either stable or serves as an “intermediate” of an active demethylation process [50]. Gene expression is commonly associated with 5hmC deposition in gene bodies [51] and differential 5hmC mapping in several cancers, including CRC has been achieved [52]. With these reports in mind, we sought to explore 5mC and 5hmC and CXCR4 upregulation in CRC. We hypothesized that enhanced CXCR4 gene expression in CRC could be the result of epigenetic modifications, either through demethylation of the CXCR4 promoter or an increase in 5hmC modification in the CXCR4 gene body.
While past studies utilized immunohistochemistry in determining CXCR4 protein expression in human CRC, we combined proteomic and transcriptomic approaches of Western blotting and RT-PCR with 5mC combined bisulfite restriction analysis (COBRA) [53] to measure CXCR4 protein and mRNA expression along with promoter methylation status in CRC tumors. Moreover, for the first time, normal stromal cells and tumor-associated stromal cells were investigated for CXCR4 mRNA and protein expression. In addition, CXCR4 analyses using publicly available databases of CRC transcriptomes, methylomes and 5-hydroxymethylomes were conducted to address our hypothesis. Herein, for the first time, we report that while 5mC distribution in the CXCR4 promoter was not significantly changed in DNA from primary colon cancer tissue (compared to control tissue DNA), we observed that enhanced CXCR4 expression in CRC associates with increased 5hmC deposition in the gene body.
2. Results
2.1. CXCR4 Is Highly-Expressed in Colorectal Cancers
A prior study reported CXCR4 protein was upregulated in CRC compared to colonic mucosa and colonic adenomas [8]. In other studies, CXCR4 mRNA and protein were shown to be upregulated in metastatic tissue of the liver and lymph nodes compared to primary CRC tumors [30,37]. However, in the prior studies, it was not determined if CXCR4 overexpression correlated with CXCR4 in tumor colonocytes and/or in tumor stromal cells. Therefore, to extend our understanding of CXCR4 regulation in colorectal cancer, we separated colonocytes from stromal cells and examined their CXCR4 expression levels compared to their control counterparts.
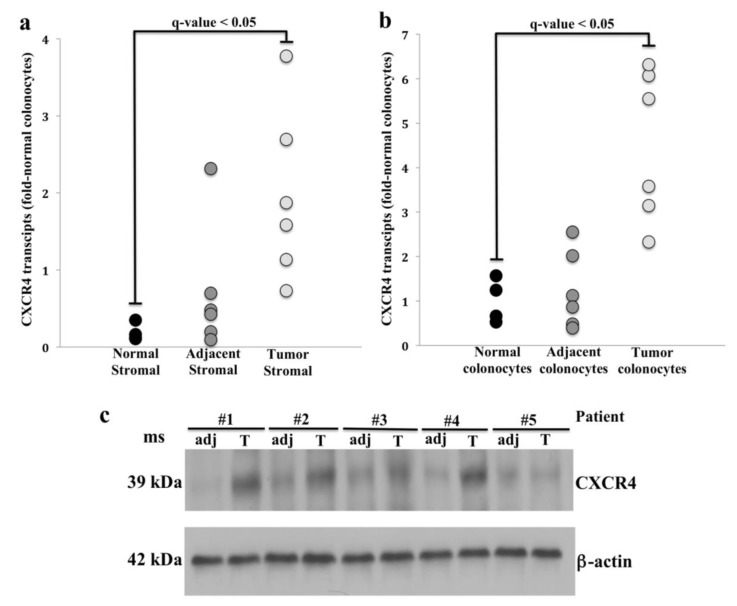
We first examined CXCR4 mRNA levels in colon tissues from four healthy controls and six CRC patients. We found in each case that colonocytes from normal colon, adjacent colon and cancers more strongly expressed CXCR4 compared to stromal cells matched to the same tissue (q-value < 0.05) (Figure 1a,b). While CXCR4 expression was lower in stromal cells, tumor-associated stromal cells expressed CXCR4 more strongly than normal stromal cells (Figure 1a).
Figure 1.
C-X-C motif chemokine receptor 4 (CXCR4) transcript and protein expression in tumor and adjacent normal tissue. RT-PCR analysis of CXCR4 transcript abundance in normal and tumor tissue: (a) stromal cells and (b) colonocytes after multiple testing correction (Tukey post hoc test, Supplementary Table S1). Note the increase of CXCR4 transcripts in both tumor stromal cells and colonocytes compared to adjacent matched tissue controls (q-value < 0.05). (c) CXCR4 protein expression in tumor colonocytes (T) and normal colonocytes adjacent to tumor (adj). Note the increased CXCR4 protein expression in tumor colonocytes compared to adjacent colonocytes from normal appearing mucosa (n = 5; p-value < 0.05, paired Student’s t-test, Supplementary Table S2).
We next evaluated CXCR4 protein expression in tumor colonocytes from primary freshly isolated colorectal tumors and colonocytes from matched adjacent normal-appearing colonic mucosa. Overall, CXCR4 protein expression correlated with qPCR findings with increased CXCR4 protein in tumor colonocytes compared to colonocytes from adjacent mucosa (Figure 1c). To assess the effects of cancer stage on CXCR4 expression, we analyzed RNA-seq data in n = 1625 CRC patients from the TCGA. Normalized CXCR4 transcript reads (FPKM values) served as the basis for comparing mRNA expression differences among the CRC that were categorized into 4 tumor stages. We found that there was a wide variation in the FPKM distribution within each tumor stage, ranging from low expression (FPKM value = 1) to high expression (FPKM value = 61). However, when comparing mean FPKM values, we did not find statistically significant differences among tumor stages (Supplementary Figure S1).
2.2. CXCR4 Promoter Hypomethylation Is Associated with Metastatic CRC
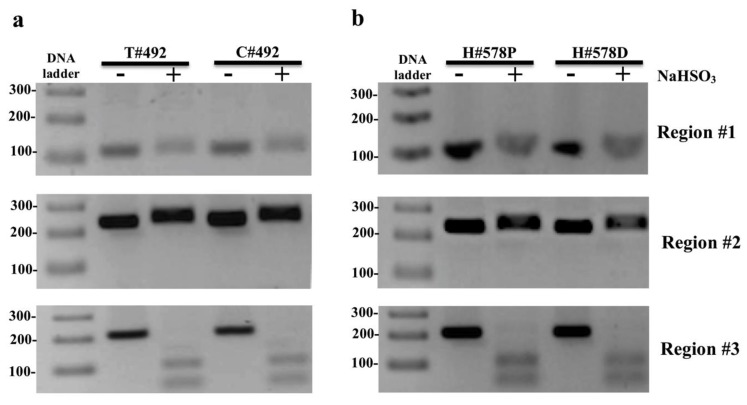
To investigate 5mC methylation in CXCR4 in human colon cancers, we extracted DNA from tumor colonocytes and adjacent normal-appearing colonocytes and tested DNA for the presence of 5mC in 3 CXCR4 genomic loci by COBRA as illustrated in Supplementary Figure S2. Each sample was run on two lanes: lane (1) bisulfite-treated PCR amplified DNA without restriction enzyme digestion, which served as a reference control for unmethylated CpG and lane (2) bisulfite-treated PCR amplified DNA digested with a restriction enzyme recognizing amplicons containing a 5’mCpG sequence. Amplicons of region #3 showed complete digestion indicating methylation of tumor and adjacent control samples (Figure 2a).
Figure 2.
Representational combined bisulfite restriction analysis (COBRA) results of 3 regions of interest located in CXCR4 from one colorectal cancer (CRC) patient: (a) (#492) and one healthy individual (Supplementary Figure S3) (b) (#578) (Supplementary Figure S4). Each sample was run on two lanes: lane 1) bisulfite-treated PCR amplified DNA without restriction enzyme digestion, which served as a reference control for unmethylated CpG and lane 2) bisulfite-treated PCR amplified DNA digested with a restriction enzyme recognizing amplicons containing a 5’mCpG sequence. Region #3 shows complete PCR product digestion, indicating that region #3 possesses cytosine methylation (5mC) CXCR4 in tumor colonocytes (T# patient ID number), adjacent normal colonocytes (C# patient ID number), and proximal/distal colon of healthy individuals (H# patient ID number P/D proximal/distal colon). Restriction enzyme digestion was not evident in regions #1 or #2 in any tumor or normal colonocytes from any CRC patient or controls and therefore unmethylated in both regions. PCR product treated with restriction enzyme lane = +; PCR product from untreated sample lane = −; Restriction enzyme sensitive digestion was not detected for the amplicons of the intragenic CpG island region #2 or promoter region #1 in either tumor or control samples, indicating the absence of 5mC in those regions (Figure 2a). In addition to CRC samples, regions #1–#3 were assessed in colonocytes from proximal and distal colon in five healthy control individuals. The 5mC results of CXCR4 for colonocytes from healthy control individuals were identical to those of malignant colonocytes from CRC patients, with 5mC present in region #3, but absent in regions #1 or #2 (Figure 2b). Whole COBRA gel images of region #1–#3 for the other CRC and normal control colons tested for CXCR4 5mC are shown in Supplementary Figures S3 and S4, respectively.
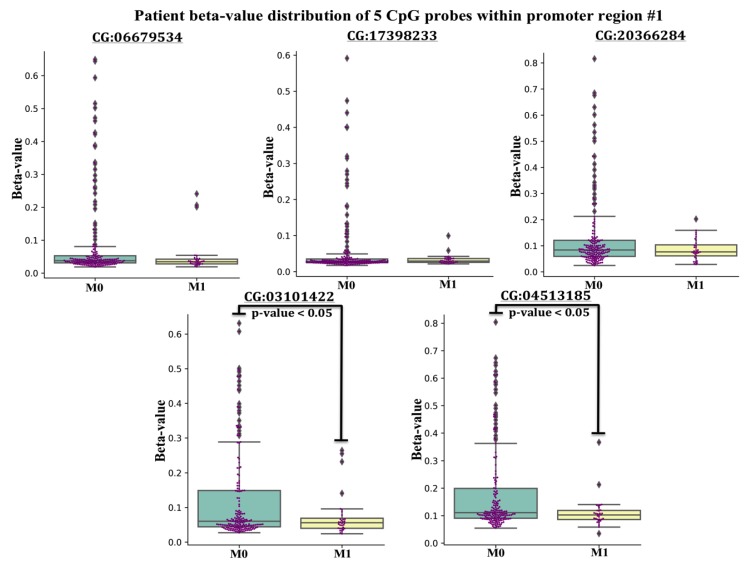
The lack of 5mC in regions #1 and #2 in colorectal cancers, prompted an additional study to evaluate the 5mC beta-value (methylation) distribution of 5’mCpG probes (n = 5) located within promoter region #1 across n = 371 tumors from CRC patients archived in TCGA that were classified by the Tumor, Node, Metastasis (TNM) staging system (M0 = no evidence of metastasis, M1 = evidence of metastasis). Typically, normalized 450K 5mC beta-values of CpG probes ranging from 0.00–0.19 are interpreted as unmethylated, whereas beta values ranging from 0.20–0.59 are considered partially methylated, and beta values > 0.59 are considered fully methylated [54]. The beta-distribution of CpG probes located within promoter region #1 revealed three findings. First, the vast majority of beta-values of each CpG probe were under 0.20, indicating that the promoter region #1 of CXCR4 was unmethylated in CRC in agreement with our COBRA findings (Figure 3).
Figure 3.
5 Boxplots displaying methylation beta-values (Y-axis) of 5 probes within promoter CpG island region #1, extracted from CRC patients classified in the Tumor, Node, Metastasis (TNM) staging system as M1 = evidence of metastasis and M0 = no evidence of metastasis from the TCGA registry. The [CG########] ID on the X-axis of each boxplot represents the unique ID of each probe within the 450K-array data set. A student t-test was conducted in finding mean beta-value differences for two CG probes between M0 and M1 cohorts (p-value < 0.05).
Since the vast number of CRC have low beta values in the promoter region #1, it is not surprising that we did not detect 5mC in region #1 in tumors from our patient subset. Secondly, some tumors from CRC patients had beta-values of CpG probes exceeding the 0.19 threshold, suggesting region #1 of the CXCR4 promoter region #1 was partially methylated in a subset of CRC. Lastly, the average beta-value for 2 of 5 CpG probes was statistically lower in the metastatic (M1) cohort compared to patients without metastasis (M0), suggesting that CXCR4 promoter hypomethylation may be occurring in metastatic CRC (Figure 3). Taken together, this suggests that hypomethylation of the CXCR4 promoter region #1 associates with more aggressive CRC.
2.3. CXCR4 Promoter Hypermethylation Correlates with Decreased Expression in CRC Cell Lines
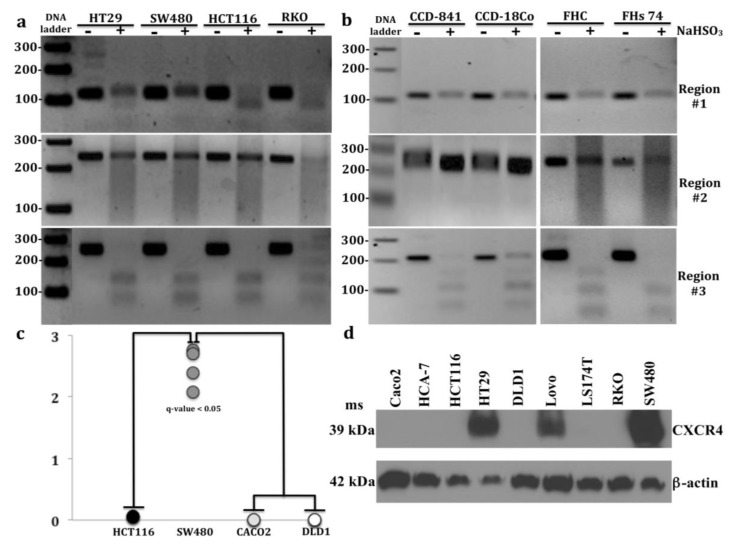
In addition to tumors and adjacent normal colonic mucosa, we investigated CRC cell lines, HT29, SW480, HCT116, and RKO colonocytes; and untransformed colon cell lines, FHC, FHs 74, and CCD-841 colonocytes and CCD-18Co fibroblasts for the presence of 5mC in regions #1-#3. Consistent with results in primary tissues, region #3 was fully methylated in 3 of 4 CRC cell lines, HT29, SW480, and HCT116 cells and in 2 of 4 untransformed colon cell lines, FHC and FHs-74 cells (Figure 4a,b).
Figure 4.
Combined bisulfite restriction analysis (COBRA) results of 3 regions of interest located in CXCR4 from: (a) 4 CRC cell lines: HT29, SW480, HCT116, and RKO cells and (b) 4 untransformed colon cell lines: CCD-841, CCD-18Co, FHC, and FHs 74 cells. Region #3 shows complete PCR product digestion in CRC cell lines: HT29, SW480, and HCT116 cells and untransformed cell lines: FHC and FHs 74 cells, indicating that region #3 is fully methylated. Incomplete PCR product digestion in RKO cells and untransformed colon cell lines: CCD-841 and CCD-18Co cells indicate partial methylation in region #1. Incomplete PCR digestion was only evident in RKO cells in region #2 indicating partial methylation. 3 of 4 CRC cell lines: HT29, HCT116, and RKO cells displayed incomplete PCR digestion in region #1, whereas no PCR digestion was evident in any untransformed colon cell line cells. (c) RT-PCR analysis of CXCR4 transcript abundance in 4 CRC cell lines: HCT116, SW480, Caco2, and DLD1 cells (q-value < 0.05) after multiple testing correction (Tukey post hoc test, Supplementary Table S1). Note the increase of CXCR4 transcripts in SW480 cells compared to HCT116, Caco2, and DLD1 cells. (d) Western blot analysis of CXCR4 in 9 CRC cell lines: Caco2, HCA-7, HCT116, HT29, DLD1, Lovo, LS174T, RKO, and SW480 cells (Supplementary Table S2). PCR product treated with restriction enzyme lane = +; PCR product untreated lane = −.
However, as we found only partial amplicon digestion for region #3 in CRC cell line RKO and non-transformed colon cell lines CCD-841 and CCD-18Co, we concluded that region #3 of these cell lines was only partially methylated.
Interestingly, CRC cell line RKO was the only cell line that exhibited partial methylation in region #2. Furthermore, differential methylation between CRC and untransformed colon cell lines were more pronounced in promoter region #1, where 3 of 4 CRC cell lines: HT29, HCT116, and RKO were partially methylated, whereas none of the 4 untransformed colon cell lines were methylated. Therefore, it is reasonable to suggest that methylation within promoter region #1 may be associated with CXCR4 silencing or downregulation in these 3 CRC cell lines. Increased 5mC in region #1 and little or no transcript or protein was detected in HCT116 cells and RKO cells (Figure 4c,d).
Conversely, low 5mC was detected in HT29 cells, where CXCR4 protein was expressed. Only 3 of 9 CRC cell lines had high protein expression of CXCR4: HT29, Lovo, and SW480 cells, with strongest expression in SW480 cells. Given that CXCR4 upregulation correlates with CRC metastasis and SW480 and Lovo cells possess strong migratory properties, high CXCR4 protein expression may well contribute to their migratory properties [55]. In contrast, CRC cell lines: Caco2, HCT116, and DLD1 had undetectable CXCR4 protein levels (Figure 4d) and very low CXCR4 transcript levels (Figure 4c). Surprisingly, HT29 cells exhibited both CXCR4 protein expression and promoter methylation.
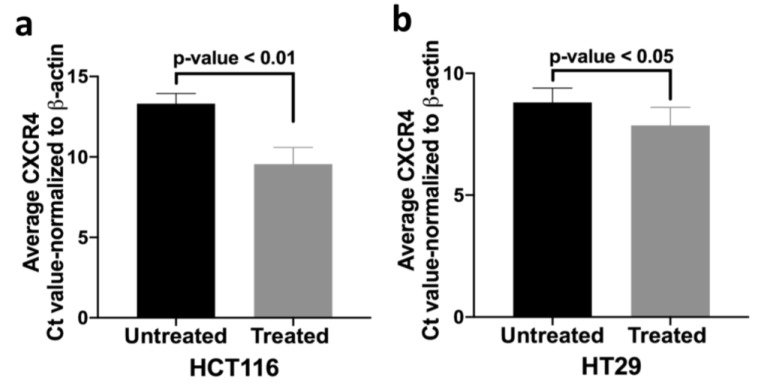
We therefore investigated whether promoter methylation might modulate CXCR4 expression by administering a demethylating agent (5-aza-2’-deoxycytidine) to HCT116 and HT29 cells. After 5-aza treatment there was increased CXCR4 mRNA compared to untreated cells (Figure 5). These results suggest that promoter hypermethylation might partially inhibit CXCR4 mRNA expression in HCT116 and HT29 cells. We speculate that increased 5mC in CXCR4 promoter might similarly inhibit CXCR4 expression in some tumors and control colons.
Figure 5.
Demethylating agent 5-aza-2’deoxycytidine increases CXCR4 transcript abundance. RT-PCR results display a decreased normalized Ct value (average) and therefore reflects the increased CXCR4 mRNA expression in colorectal cancer cell lines (a) HCT116 and (b) HT29 after treatment with 5 µM concentration of demethylating agent 5-aza-2’-deoxycytidine.
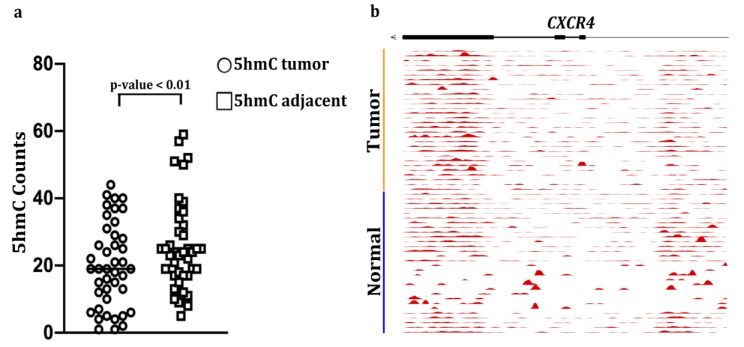
2.4. Increased 5hmC Deposition within CXCR4 Gene Body Correlates with Increased CXCR4 Expression
Since we did not detect any relationship between 5mC levels and CXCR4 expression in primary CRC vs. controls, we next examined levels of another epigenetic mark, 5hmC, in CXCR4. This epigenetic modification is frequently associated with active gene transcription [56]. We found that 5hmC was increased significantly in the CXCR4 gene bodies in n = 42 cases of CRC compared to matched adjacent normal-appearing colonic mucosa (Figure 6a). In our CRC cohort 5hmC accumulation was most abundant within the terminal 3’ end of CXCR4 (Figure 6b). This phenomenon has been reported in several genome-wide studies [56,57,58,59]. For example, a study by Lin et al., found that the average 5hmC read counts were consistently higher towards the 3’ end for many genes expressed in brain and liver tissues [56]. The authors speculated that a bias for 5hmC modification might begin near the end of the ORF of these genes. The underling mechanism for this bias, however, will require further study. Our observation of increased 5hmC in the CXCR4 gene body supports the hypothesis that 5hmC modification associates with increased CXCR4 expression.
Figure 6.
5-hydroxymethylcytosine (5hmC) abundance in CXCR4 gene bodies in colon cancers and matched adjacent mucosa (n = 42 samples, p < 0.01, paired Student’s t-test): (a) Counts per million reads at CXCR4 gene, (b) plus promoter (upstream 3kb region) in matched tumors and adjacent healthy tissue in 30 colorectal cancer patients. The moving averages at 0.01 smoother span are shown. black bars mark exons.
3. Discussion
CXCR4 is deregulated in several cancer types and appears to be important in CRC metastasis as suggested by increased CXCR4 mRNA and protein in liver [30,37] and increased protein in lymph nodes [30] in metastatic tumors compared to primary tumors. This is further supported by increased metastatic properties in colon cancer cells following CXCR4 activation and inhibition of these characteristics with anti-CXCR4 antibodies [8]. Our studies are the first to have measured CXCR4 in matched isolated malignant colonocytes and tumor-associated stroma by Western blotting. For the first time, we also observed that CXCR4 mRNA expression levels were higher in tumor-associated stromal cells compared to control stromal cells, suggesting that CXCR4 may play a critical role in regulating the tumor microenvironment as observed for other genes that are highly expressed in CRC-associated stromal cells [60,61]. The expression patterns of CXCR4 in our study were consistent with prior reports that analyzed CXCR4 expression in CRC. For example, RNA-seq expression profiles from a large sample of colon tumors in TCGA, revealed that CXCR4 mRNA expression in tumors remained stable across tumor stages (I–IV). This result was consistent with two earlier studies that compared CXCR4 mRNA expression among CRC tumor stages [30,37]. In addition, our Western blotting results showed increased CXCR4 expression in tumor colonocytes compared to matched adjacent normal-appearing colonocytes, and were in agreement with Ottaiano and colleagues’ findings of increased CXCR4 in malignant colonocytes as assessed by immunostaining [8].
Of the cell lines examined in the report, N = 6 of 9 CRC cell lines: RKO, HCT116, LS174T, Lovo, DLD1, and HCA-7 cells exhibit a hypermutable phenotype and are classified as micro-satellite instable (MSI) tumor cell lines, whereas the remaining three CRC cell lines: HT29, SW480, and Caco2 cells are classified as microsatellite stable (MSS) tumor cell lines. The microsatellite unstable phenotype is associated with poor overall survival and is caused by biallelic loss of a DNA mismatch repair enzyme, which accounts for about 15% of all sporadic CRCs [62,63]. One study concluded that overall most genes were downregulated in MSI tumors, but noted upregulation of mucin-related genes as a key molecular biomarker characterizing MSI tumors [64]. Interestingly, CXCR4 protein expression in microsatellite unstable CRC cell lines was detected only in Lovo cells, which represents 1 of 6 (17%) MSI tumors assayed; but CXCR4 was detected in 2 of 3 (67%) of MSS tumors assayed: HT29, SW480 cells. Further studies comparing primary MSI and MSS tumors will be needed to determine whether CXCR4 expression levels differ between these tumor types.
To examine potential epigenetic mechanisms regulating CXCR4 in CRC, we assessed 5mC modifications in CXCR4 by COBRA analysis, TCGA methylation data, and 5hmC modifications by publicly available nano-hmC-Seal data. The 5mC portion of the study was initially prompted by the fact that the 450 K methylation data from the UCSC Genome Browser identified CXCR4 promoter hypermethylation in HCT116 and Caco-2 cells, which lacked CXCR4 protein expression. In addition, the 5mC pattern spanning CXCR4 in HCT116 and Caco-2 cells served as a template in identifying potential regulatory hotspots to test for the presence/absence of 5mC in CRCs. In turn, regions #1 and #2 were identified as both regions overlap a CpG island within the promoter or a 5’ regulatory region in CXCR4 and are therefore most likely to impact the transcription status of CXCR4. The COBRA technique was chosen in that it allowed for a greater number CpGs to be evaluated in regions #1 and #2 (n = 9) compared to other forms of methylation-interrogating end-point PCR techniques like methylation-specific PCR (MSP).
In our sample set of CRC tissues we did not observe decreased 5mC in CXCR4 despite increased mRNA and protein expression in these tumors. Upon analysis of CRC tumor methylation in the TCGA, we found that promoter CXCR4 5mC hypermethylation exists in a small subset of primary colon cancers and those primary tumors with metastasis exhibit loss of 5mC in the promoter region. Recent studies of TCGA for both DNA 5’mCpG methylation and RNA-seq analysis suggest that 5’mCpG methylation could exert gene-specific positive or negative effects on gene transcription [65,66]. We also found increases in 5hmC in the CXCR4 gene body in an independent cohort of colon tumors [52], consistent with 5hmC serving as a marker of active gene transcription. A limitation of our 5hmC analysis is that expression levels of CXCR4 mRNA and protein were not available for this cohort and the TCGA cohort did not have 5hmC analysis.
Although, the mechanisms underlying aberrant methylation patterns in cancer remain incompletely understood, global hypomethylation, associated with genomic instability and promoter hypermethylation of tumor suppressor genes are observed in many tumors, including colon cancer [41,42,43,44,45,46]. The latter may be related to increased enzymatic activity of DNA methyltransferases in CRC compared to normal colonic mucosa [67]. Whether changes in 5hmC and/or 5mC promoter methylation regulate CXCR4 expression in metastatic CRC will require additional studies.
In CRC cell lines our results suggest that 5mC present within CpG island region #1 of the promoter may contribute to CXCR4 gene silencing in HCT116 and RKO cells as negligible levels of CXCR4 mRNA and no protein were detected in HCT116 cells and no CXCR4 protein was detected in RKO cells. Indeed, our COBRA findings of 5mC located in the CpG rich region #1 of the CXCR4 promoter in HCT116 cells are consistent with CXCR4 promoter methylation data in HCT116 cells available in the UCSC Genome Browser. While CXCR4 was still expressed in HT29 cells, despite CXCR4 promoter methylation, our demethylation studies suggest that DNA methylation limits CXCR4 expression in these cells. In this regard, treating both HT29 and HCT116 cells with DNA hypomethylating agent (5-aza) increased CXCR4 mRNA. A previous study in MCF7 breast cancer cells also demonstrated that 5-aza treatment demethylated the CXCR4 promoter and concomitantly increased CXCR4 mRNA expression [68]. We speculate that similar demethylation-induced changes in the CXCR4 promoter are occurring in (5-aza) treated HT29 and HCT116 cells that showed increased CXCR4 mRNA. Persistent CXCR4 gene expression in HT29 cells might result from lower levels of 5mC methylation in region #1 or in other CXCR4 promoter regions compared to those of HCT116 and RKO cells. We were only able to interrogate one CpG site in region #1 and in the promoter region, because of design limitations of suitable primers, thereby limiting the number of promoter associated CpG islands tested in this study. Future studies will also address potential differences in 5hmC in the CXCR4 promoters in these cell lines that could also contribute to differences in CXCR4 expression
4. Materials and Methods
4.1. Human Tissue Collection
Human colonic adenocarcinomas and adjacent normal appearing colonic mucosa were obtained for the Department of Surgical Pathology at The University of Chicago under an approved IRB protocol (10-209-A). Resected tissues were placed in an ice bath and transported promptly to the Surgical Pathology Department. Representative tumor sections and adjacent normal appearing colonic mucosa were dissected free of underlying muscle. Care was taken to avoid areas with visible necrosis. Colonocytes and stromal cells were isolated as previously described and purities assessed by cytokeratin20 (colonocyte marker) and vimentin (stromal cell marker) Western blotting [69]. Normal colon tissues were obtained from colon biopsies taken 20 cm from the anus under the approved IRB 10-209-A. For DNA methylation studies, biopsies were stored in RNA later that also preserves DNA.
4.2. Isolation of Coloncytes and Stromal Cells from Colonic Mucosa
Colonocytes and stromal cells were isolated following previously published methods with minor modifications [70,71,72]. Briefly, colon tissues were removed and mucosa scrape-isolated and minced with razor blade into 2 mm fragments that were collected in tubes containing 6 mL sterile ice-cold transport media, 50 IU/mL penicillin (Millipore Sigma, St. Louis, MO, USA), 50 µg/mL streptomycin (Millipore Sigma, St. Louis, MO, USA) and 0.5 mg/mL gentamycin (Millipore Sigma, St. Louis, MO, USA). Tissue was washed 3 times by gentle inversion and collected by gravity sedimentation and re-suspended in 10 mL chelating buffer (transport media plus 1 mM EDTA (Millipore Sigma, St. Louis, MO, USA) and 1 mM EGTA (Millipore Sigma, St. Louis, MO, USA). Tissue was incubated on a shaker overnight at 4 °C to release colonocytes into the supernatant. The pellet was washed three times with 3 mL ice-cold PBS (Thermo Fisher Scientific, Waltham, MA, USA), releasing residual colonocytes, and colonocyte-containing supernatants were combined. Supernatants containing the epithelial cell fraction, and pellets containing the stromal cell fraction were centrifuged at 400× g for 5 min at 4 °C, yielding loosely packed pellets from which proteins were solubilized in 2× Laemmli buffer (Millipore Sigma, St. Louis, MO, USA).
4.3. Cell Culture
All colon cancer and normal colon cell lines were obtained from ATCC and cultured at 37 °C in a humified atmosphere of 95% O2 and 5% CO2 and 10% fetal calf serum (Thermo Fisher Scientific, Waltham, MA, USA). Cell authentications for all lines were confirmed by short tandem repeats by (IDEXX Bioanalytics, Columbia, MO, USA). Cells were shown to be mycoplasma free by IDEXX. HCT116 and FHC cells were grown in high glucose DMEM medium (Thermo Fisher Scientific, Waltham, MA, USA). DLD-1 cells were cultured in RPMI-1640 media (ATCC, Manassas, VA, USA). SW480 cells were grown in ATCC-formulated Leibovitz’s L-15 Medium (ATCC, Manassas, VA, USA). RKO, LS-174T, Caco2, CCD-841, CCD-18Co cells were grown in ATCC-formulated Eagle’s Minimum Essential Medium (ATCC, Manassas, VA, USA). Lovo cells were cultures in ATCC-formulated F-12K Medium (ATCC, Manassas, VA, USA). FHs 74 cells were grown in ATCC-formulated Hybri-Care medium (ATCC, Manassas, VA, USA). HCA-7 cells, derived from a rectal carcinoma {Marsh, 1993 #1850}, were generously provided by Dr. Susan Kirkland (ICRF, London UK). Cells were grown in McCoy’s media (ATCC, Manassas, VA, USA).
4.4. Measurement of CXCR4 Steady-State Transcript Levels
mRNA was extracted from colonocytes and stromal cells isolated from tumors and control colon (CRC n = 6; control colons n = 4) and from CRC cell lines: HCT116, SW480, Caco2, DLD1 and cells using the RNeasy® Mini Kit (Qiagen, Germantown, MD, USA). mRNA (150 ng) was then reverse transcribed into cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Beverly, MD, USA) in 20 μL total volume. Incubation conditions were 37 °C for 60 min, and 95 °C for 5 min. The resulting first-strand cDNA was used as template for quantitative PCR in triplicate using Fast SYBR Green QPCR Master Mix kit (Applied Biosystems, Beverly, MD, USA). Reverse transcribed cDNA (1:10 dilution) and primers were mixed in 20 µL with fast SYBR green master mixture. Reactions were monitored on the LightCycler® 480 II System (Roche Diagnostic Corporation, Basel, Switzerland). Reactants were initially heated to 95 °C for 20 s followed by 40 cycles: denaturation step at 95 °C for 10 s, annealing step at 55 °C for 15 s and extension step at 60 °C for 30 s. PCR amplification was verified by melting curve and electrophoretic analyses of the PCR products on a 2% agarose gel. Negative control reactions were also included (omitting reverse transcriptase or template cDNA). Data were analyzed using the comparative 2^(-ΔΔCt) method [73] and mRNA levels were normalized to β-actin and expressed as fold-control (Supplementary Table S1). The multiple comparisons analysis using Tukey’s method (q < 0.05) consisted of 11 groups, including: human whole tissue, control colonocytes and stromal cells, colonocytes and stromal cells from normal appearing mucosa adjacent to tumor, tumor colonocytes and tumor stromal cells. CXCR4 expression was also assessed in 4 CRC: HCT116, SW480, Caco2, and DLD1 cell lines. Primer sequences for quantitative RT-PCR include:
β-actin Forward primer (5’->3’) CAGCCATGTCGTTGCTATCCAGG;
β-actin Reverse primer (5’->3’) AGGTCCAGACGCAGGATGGCATG;
CXCR4 Forward primer (5’->3’) GCGGTTACCATGGAGGGGAT;
CXCR4 Reverse primer (5’->3’) CCCATGACCAGGATGACCAAT;
4.5. CXCR4 Western Blotting
Western blotting was performed to assess CXCR4 expression in primary colon cancers and adjacent normal appearing colonic mucosa, and 9 colorectal cell lines: Caco2, HCT116, HCA-7, HT29, DLD1, Lovo, LS174T, RKO, and SW480 colonocytes. Proteins were extracted in SDS-containing Laemmli buffer, quantified by RC-DC protein assay and subjected to Western blotting. Sample lysates (60 μg protein) and pre-stained molecular weight markers were separated by SDS-PAGE on 4%–20% resolving polyacrylamide gradient gels and electroblotted to PVDF membranes. Blots were incubated overnight at 4 °C with specific primary rabbit monoclonal antibody at 1:1000 dilution (CXCR4, Abcam ab181020, Cambridge, MA, USA) followed by 1hr incubation with appropriate peroxidase-coupled secondary antibodies. CXCR4 Abcam Cat#ab181020 Blocking: 5% milk for 3 h 1°AB 1:1000 in 5% milk overnight at 4 °C secondary AB 1:3000 anti-rabbit antibodies in 5% milk for 1 h at RT that were detected by enhanced chemiluminescence using X-OMAT film (Carestream, Rochester, NY, USA). Xerograms were digitized using Epson Perfection V600I scanner (Epson, Long Beach, CA, USA). and CXCR4 band intensity was quantified using Un-Scan-It software V6.3 (Silk Scientific Inc., Orem, UT, USA) [69] (Supplementary Table S2). A paired t-test was conducted to determine CXCR4 band intensity differences between matched CRC tumor and adjacent tissue samples.
4.6. Combined Bisulfite Restriction Analysis
Prior to COBRA [53], genomic DNA (gDNA) was extracted and eluted in water from human tissue samples and colorectal cell lines (HCT116, RKO, HT29, and SW480) using the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD, USA). Clinical information for patients for the methylation studies is summarized in Supplementary Table S3. Briefly, gDNA (~200 ng) was treated with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo, Irvine, CA, USA) per the manufacturer’s protocol. Bisulfite-treated DNA was eluted in water. Bisulfite converted DNA (~200 ng) of bisulfite converted DNA was used as template under standard thermal conditions (15 min hot start (94 °C denaturation for 30 s, 56 °C annealing temp, and 72 °C extension for 40 cycles) 72 °C final extension for 10 min) in a 25 μL reaction using the PyroMark PCR kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. The bisulfite-specific primers with sequences designed by MethPrimer software [74] include:
Region #1: Promoter CpG island upstream of CXCR4 TSS [1 CpG] HpyCH4IV
Forward primer: (5’>3’)TTTTGAGTAGAGGATAAGTTTTGGTA;
Reverse primer: (5’->3’) ACAAACCTCAACCAATCTAAAATC;
Region #2: Intragenic CpG island at the 5’ regulatory end of CXCR4 [8 CpGs] BstUI
Forward primer: (5’>3’)GGTGGTTATTGGAGTATTTAGGTTTT;
Reverse primer: (5’->3’)AACAAAATCCCTAAACTTC;
Region #3: Terminal 3’ exon of CXCR4 [2CpGs] HpyCH4IV
Forward primer: (5’->3’)TTATTTATAAGTTATTGGGGTAGAAG;
Reverse primer: (5’->3’)ATCCTAACCTTCATCAATCTAAAC;
PCR products (~7 μL) were digested in a 25 μL reaction containing either BstUI (60 °C) or HpyCH4IV (37 °C) restriction enzyme for 4 h. Restriction digests were then resolved on a 2.5% agarose gel with ethidium bromide. BstUI: cuts 5’CGCG’3 sequence and HpyCH4IV: cuts 5’ACGT’3 sequence.
4.7. 5hmC-Modified Locations in the CXCR4 Gene Body
5hmC-Seal data were downloaded for 42 pairs of tumor (TU) and adjacent tissue (TI) samples from patients with colorectal cancer (GSE89570) [52]. Raw sequencing reads were summarized for the gene bodies or promoters according to the current GENCODE annotations (release 19). The normalized counts from DESeq2 by library size were log2 transformed and corrected for batch effect using linear regression. The paired t test was used to evaluate whether the 5hmC modification levels in CXCR4 gene body were different between tumors and adjacent tissues (p-value < 0.01).
4.8. Inhibition of 5mC Deposition in Colorectal Cancer Cell Lines HCT116 and HT29
HCT116 and HT29 cells were seeded at ~10,000 cells at day -2. On day 0, culture medium was removed and replaced with fresh medium containing 5µM 5’-aza-2’-deoxycytidine (Millipore Sigma, St. Louis, MO, USA) dissolved in DMSO and incubated for 72 h. On day 3, cells were rinsed twice with PBS and harvested for RNA and proteins. CXCR4 mRNA expression was analyzed by RT-PCR as previously described.
4.9. CXCR4 Bioinformatics Analysis
High-throughput data from tumors from CRC patients were extracted from The Cancer Genome Atlas (TCGA) and utilized for dissecting relationships among 5mC methylation, mRNA gene expression and clinical correlates [75]. CRC were categorized into respective TNM (M0, M1) or tumor stages (I–IV) for analysis. A python statistical data visualization library (Seaborn) was used to generate boxplots showing mRNA expression and methylation distributions [76]. The x-axis represents the TNM staging classification of CRC and the y-axis represents FPKM (mRNA gene expression) values from RNA-seq data or beta (methylation) values from 450K methylation microarray probes spanning two CpG islands within CXCR4 (Supplementary Table S4).
5. Conclusions
In summary, we confirmed upregulation of CXCR4 in CRC and extended prior studies to show by cell-specific isolation and Western blotting that malignant colonocytes in CRC have the highest expression levels. Furthermore, for the first time, we demonstrate that tumor-associated stromal cells express increased CXCR4 mRNA. These changes in CXCR4 expression were not accompanied by changes in 5mC, but rather by previously unreported increased 5hmC that we postulate marks this gene for increased transcription in CRC. Multiple mechanisms regulating CXCR4 expression in CRC are likely involved, including DNA epigenetic processes such as increases in 5hmC in the CXCR4 gene body [52], acquisition of activating and loss of repressive histone marks such as increased H3K4me3 and decreased H3K27me3, respectively, [77]; and recruitment of trans-activating factors such as nuclear respiratory factor1 (NRF1) [78]. Additional studies to discover new or novel genetic and epigenetic mechanisms regulating CXCR4 are warranted to potentially target CXCR4 in colon cancer.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/3/539/s1, Figure S1: Normalized CXCR4 FPKM values from CRC patients categorized by tumor stage, Figure S2: UCSC Genome Browser (hg19 build) depiction of CXCR4 containing 2 CpG islands (green) and 3 Methyl 450K probe sub tracks for colorectal cell lines Caco2 and HCT116 (Probe colors of Blue = unmethylated, Purple = partially methylated, and Yellow = fully methylated), Figure S3: Full-length COBRA images from Figure 2a (T tumor, C control), Figure S4: Full-length COBRA images from healthy control patients from Figure 2b (H# human subject number; P proximal colon, D distal colon), Table S1: Clinical characteristics and statistical analysis of patients used in the qPCR studies, Table S2: Densitometry statistics of CXCR4 bands in Figure 1c and Figure 4d. Table S3: Clinical characteristics of patients used in the COBRA analysis in Figure 2 and Figures S3 and S4. Table S4: Number of patients deposited in TCGA that were used in the CXCR4 RNA-seq and 450K methylation microarray analysis.
Author Contributions
All authors have read and agree to the published version of the manuscript. Conceptualization, A.J.S., M.B., and S.K.; Methodology, A.J.S., S.Z., Q.Z., U.D., R.M., and T.K.; Validation, A.J.S., Q.Z.; Formal Analysis, A.J.S., W.Z., S.Z. and X.Z.; Investigation, A.J.S., M.B., and S.K.; Writing—Original Draft Preparation, A.J.S. and M.B.; Writing—Review and Editing, W.Z., D.C.W.-S., T.J., E.P., M.B., and S.K.
Funding
This research was funded by Veterans Affairs 2I01BX000824 (SK), National Cancer Institute, grant numbers 1RO3CA216160 (SK), 1R01CA240710 (MB) and The University of Missouri, School of Medicine Bridge Funding (SK).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Burger J.A., Kipps T.J. CXCR4: A key receptor in the crosstalk between tumor cells and their microenvironment. Blood. 2006;107:1761–1767. doi: 10.1182/blood-2005-08-3182. [DOI] [PubMed] [Google Scholar]
- 2.Wojcechowskyj J.A., Didigu C.A., Lee J.Y., Parrish N.F., Sinha R., Hahn B., Bushman F.D., Jensen S.T., Seeholzer S.H., Doms R.W. Quantitative phosphoproteomics reveals extensive cellular reprogramming during HIV-1 entry. Cell Host Microbe. 2013;13:613–623. doi: 10.1016/j.chom.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., Kataoka Y., Kitamura Y., Matsushima K., Yoshida N., Nishikawa S.-I., et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 4.Klein S., Abraham M., Bulvik B., Dery E., Weiss I.D., Barashi N., Abramovitch R., Wald H., Harel Y., Olam D., et al. CXCR4 Promotes Neuroblastoma Growth and Therapeutic Resistance through miR-15a/16-1–Mediated ERK and BCL2/Cyclin D1 Pathways. Cancer Res. 2017;78:1471–1483. doi: 10.1158/0008-5472.CAN-17-0454. [DOI] [PubMed] [Google Scholar]
- 5.Barbero S., Bonavia R., Bajetto A., Porcile C., Pirani P., Ravetti J.L., Zona G.L., Spaziante R., Florio T., Schettini G. Stromal Cell-derived Factor 1α Stimulates Human Glioblastoma Cell Growth through the Activation of Both Extracellular Signal-regulated Kinases 1/2 and Akt. Cancer Res. 2003;63:1969. [PubMed] [Google Scholar]
- 6.Kijima T., Maulik G., Ma P.C., Tibaldi E.V., Turner R.E., Rollins B., Sattler M., Johnson B.E., Salgia R. Regulation of cellular proliferation, cytoskeletal function, and signal transduction through CXCR4 and c-Kit in small cell lung cancer cells. Cancer Res. 2002;62:6304. [PubMed] [Google Scholar]
- 7.Thomas R.M., Kim J., Revelo-Penafiel M.P., Angel R., Dawson D.W., Lowy A.M. The chemokine receptor CXCR4 is expressed in pancreatic intraepithelial neoplasia. Gut. 2008;57:1555–1560. doi: 10.1136/gut.2007.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottaiano A., Di Palma A., Napolitano M., Pisano C., Pignata S., Tatangelo F., Botti G., Acquaviva A.M., Castello G., Ascierto P.A., et al. Inhibitory effects of anti-CXCR4 antibodies on human colon cancer cells. Cancer Immunol. Immunother. 2004;54:781–791. doi: 10.1007/s00262-004-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuo J., Wen M., Li S., Lv X., Wang L., Ai X., Lei M. Overexpression of CXCR4 promotes invasion and migration of non-small cell lung cancer via EGFR and MMP-9. Oncol. Lett. 2017;14:7513–7521. doi: 10.3892/ol.2017.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eck S.M., Côté A.L., Winkelman W.D., Brinckerhoff C.E. CXCR4 and matrix metalloproteinase-1 are elevated in breast carcinoma-associated fibroblasts and in normal mammary fibroblasts exposed to factors secreted by breast cancer cells. Mol. Cancer Res. 2009;7:1033–1044. doi: 10.1158/1541-7786.MCR-09-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orimo A., Gupta P.B., Sgroi D.C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V.J., Richardson A.L., Weinberg R.A. Stromal Fibroblasts Present in Invasive Human Breast Carcinomas Promote Tumor Growth and Angiogenesis through Elevated SDF-1/CXCL12 Secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 12.Scotton C.J., Wilson J.L., Scott K., Stamp G., Wilbanks G.D., Fricker S., Bridger G., Balkwill F.R. Multiple actions of the chemokine CXCL12 on epithelial tumor cells in human ovarian cancer. Cancer Res. 2002;62:5930–5938. [PubMed] [Google Scholar]
- 13.Koshiba T., Hosotani R., Miyamoto Y., Ida J., Tsuji S., Nakajima S., Kawaguchi M., Kobayashi H., Doi R., Hori T., et al. Expression of stromal cell-derived factor 1 and CXCR4 ligand receptor system in pancreatic cancer: A possible role for tumor progression. Clin. Cancer Res. 2000;6:3530. [PubMed] [Google Scholar]
- 14.Robledo M.M. Expression of Functional Chemokine Receptors CXCR3 and CXCR4 on Human Melanoma Cells. J. Biol. Chem. 2001;276:45098–45105. doi: 10.1074/jbc.M106912200. [DOI] [PubMed] [Google Scholar]
- 15.Bertolini F., Dell’Agnola C., Mancuso P., Rabascio C., Burlini A., Monestiroli S., Gobbi A., Pruneri G., Martinelli G. CXCR4 Neutralization, a Novel Therapeutic Approach for Non-Hodgkin’s Lymphoma. Cancer Res. 2002;62:3106–3112. [PubMed] [Google Scholar]
- 16.Corcione A., Ottonello L., Tortolina G., Facchetti P., Airoldi I., Guglielmino R., Dadati P., Truini M., Sozzani S., Dallegri F., et al. Stromal cell-derived factor-1 as a chemoattractant for follicular center lymphoma B cells. J. Natl. Cancer Inst. 2000;92:628–635. doi: 10.1093/jnci/92.8.628. [DOI] [PubMed] [Google Scholar]
- 17.Arai J., Yasukawa M., Yakushijin Y., Miyazaki T., Fujita S. Stromal cells in lymph nodes attractB-lymphoma cells via production ofstromal cell-derived factor-1. Eur. J. Haematol. 2000;64:323–332. doi: 10.1034/j.1600-0609.2000.90147.x. [DOI] [PubMed] [Google Scholar]
- 18.Hwang J.H., Hwang J.H., Chung H.K., Kim N.W., Hwang E.S., Suh J.M., Kim H., You K.H., Kwon O.-Y., Ro H.K., et al. CXC Chemokine Receptor 4 Expression and Function in Human Anaplastic Thyroid Cancer Cells. J. Clin. Endocrinol. Metab. 2003;88:408–416. doi: 10.1210/jc.2002-021381. [DOI] [PubMed] [Google Scholar]
- 19.Gu F., Ma Y., Zhang J., Qin F., Fu L. Function of Slit/Robo signaling in breast cancer. Front. Med. 2015;9:431–436. doi: 10.1007/s11684-015-0416-9. [DOI] [PubMed] [Google Scholar]
- 20.Jin Z., Zhao C., Han X., Han Y. Wnt5a promotes ewing sarcoma cell migration through upregulating CXCR4 expression. BMC Cancer. 2012;12:480. doi: 10.1186/1471-2407-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X., Wang D., Wang X., Sun S., Zhang Y., Wang S., Miao R., Xu X., Qu X.-J. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J. Exp. Clin. Cancer Res. 2019;38:32. doi: 10.1186/s13046-018-1014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rempel S.A., Dudas S., Ge S., Gutiérrez J.A. Identification and localization of the cytokine SDF1 and its receptor, CXC chemokine receptor 4, to regions of necrosis and angiogenesis in human glioblastoma. Clin. Cancer Res. 2000;6:102–111. [PubMed] [Google Scholar]
- 23.Murakami T., Maki W., Cardones A.R., Fang H., Kyi A.T., Nestle F.O., Hwang S.T. Expression of CXC chemokine receptor-4 enhances the pulmonary metastatic potential of murine B16 melanoma cells. Cancer Res. 2002;62:7328–7334. [PubMed] [Google Scholar]
- 24.Geminder H., Sagi-Assif O., Goldberg L., Meshel T., Rechavi G., Witz I.P., Ben-Baruch A. A possible role for CXCR4 and its ligand, the CXC chemokine stromal cell-derived factor-1, in the development of bone marrow metastases in neuroblastoma. J. Immunol. 2001;167:4747–4757. doi: 10.4049/jimmunol.167.8.4747. [DOI] [PubMed] [Google Scholar]
- 25.Libura J., Drukala J., Majka M., Tomescu O., Navenot J.M., Kucia M., Marquez L., Peiper S.C., Barr F.G., Janowska-Wieczorek A., et al. CXCR4–SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood. 2002;100:2597–2606. doi: 10.1182/blood-2002-01-0031. [DOI] [PubMed] [Google Scholar]
- 26.Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S., et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 27.Jung Y.-D., Shim J.-W., Park S.-J., Choi S.H., Yang K., Heo K., Park M.-T. Downregulation of UHRF1 promotes EMT via inducing CXCR4 in human cancer cells. Int. J. Oncol. 2014;46:1232–1242. doi: 10.3892/ijo.2014.2813. [DOI] [PubMed] [Google Scholar]
- 28.Kim J.-H., Shim J.-W., Eum D.-Y., Kim S.D., Choi S.H., Yang K., Heo K., Park M.-T. Downregulation of UHRF1 increases tumor malignancy by activating the CXCR4/AKT-JNK/IL-6/Snail signaling axis in hepatocellular carcinoma cells. Sci. Rep. 2017;7:2798. doi: 10.1038/s41598-017-02935-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alevizos L., Kataki A., Derventzi A., Gomatos I., Loutraris C., Gloustianou G., Manouras A., Konstadoulakis M.M., Zografos G. Breast cancer nodal metastasis correlates with tumour and lymph node methylation profiles of Caveolin-1 and CXCR4. Clin. Exp. Metastasis. 2014;31:511–520. doi: 10.1007/s10585-014-9645-6. [DOI] [PubMed] [Google Scholar]
- 30.Murakami T., Kawada K., Iwamoto M., Akagami M., Hida K., Nakanishi Y., Kanda K., Kawada M., Seno H., Taketo M., et al. The role of CXCR3 and CXCR4 in colorectal cancer metastasis. Int. J. Cancer. 2012;132:276–287. doi: 10.1002/ijc.27670. [DOI] [PubMed] [Google Scholar]
- 31.Spoo A.C., Lubbert M., Wierda W.G., Burger J.A. CXCR4 is a prognostic marker in acute myelogenous leukemia. Blood. 2006;109:786–791. doi: 10.1182/blood-2006-05-024844. [DOI] [PubMed] [Google Scholar]
- 32.Bao L., Lai Y., Liu Y., Qin Y., Zhao X.-S., Lu X., Jiang Q., Lu J., Huang X. CXCR4 is a good survival prognostic indicator in multiple myeloma patients. Leuk. Res. 2013;37:1083–1088. doi: 10.1016/j.leukres.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Goto M., Yoshida T., Yamamoto Y., Furukita Y., Inoue S., Fujiwara S., Kawakita N., Nishino T., Minato T., Yuasa Y., et al. CXCR4 Expression is Associated with Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 2015;24:832–840. doi: 10.1245/s10434-015-4974-5. [DOI] [PubMed] [Google Scholar]
- 34.Masuda T., Nakashima Y., Ando K., Yoshinaga K., Saeki H., Oki E., Morita M., Oda Y., Maehara Y. Nuclear expression of chemokine receptor CXCR4 indicates poorer prognosis in gastric cancer. Anticancer. Res. 2014;34:6397–6403. [PubMed] [Google Scholar]
- 35.D’Alterio C., Consales C., Polimeno M., Franco R., Cindolo L., Portella L., Cioffi M., Calemma R., Marra L., Claudio L., et al. Concomitant CXCR4 and CXCR7 expression predicts poor prognosis in renal cancer. Curr. Cancer Drug Targets. 2010;10:772–781. doi: 10.2174/156800910793605839. [DOI] [PubMed] [Google Scholar]
- 36.Liu H., Liu Y., Liu W., Zhang W., Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat. Commun. 2015;6:8494. doi: 10.1038/ncomms9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J., Takeuchi H., Lam S., Turner R.R., Wang H.-J., Kuo C., Foshag L., Bilchik A., Hoon D. Chemokine Receptor CXCR4 Expression in Colorectal Cancer Patients Increases the Risk for Recurrence and for Poor Survival. J. Clin. Oncol. 2005;23:2744–2753. doi: 10.1200/JCO.2005.07.078. [DOI] [PubMed] [Google Scholar]
- 38.Li Y.-P., Pang J., Bai P.-Y., Kong P., Gao S., Wang W.-D., Cui Y. Role of CXCR4 and SDF1 as prognostic factors for survival and the association with clinicopathology in colorectal cancer: A systematic meta-analysis. Tumor Boil. 2017;39 doi: 10.1177/1010428317706206. [DOI] [PubMed] [Google Scholar]
- 39.Staller P., Sulitkova J., Lisztwan J., Moch H., Oakeley E.J., Krek W. Chemokine receptor CXCR4 downregulated by von Hippel–Lindau tumour suppressor pVHL. Nature. 2003;425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 40.Bachelder R.E., Wendt M.A., Mercurio A.M. Vascular endothelial growth factor promotes breast carcinoma invasion in an autocrine manner by regulating the chemokine receptor CXCR4. Cancer Res. 2002;62:7203. [PubMed] [Google Scholar]
- 41.Hernandez-Blazque F., Habib M., Dumollard J.-M., Barthélémy C., Benchaib M., De Capoa A., Niveleau A. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–693. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmer F., Brinkman A., Assenov Y., Matarese F., Kaan A., Sabatino L., Villanueva A., Huertas D., Esteller M., Lengauer T., et al. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics. 2012;7:1355–1367. doi: 10.4161/epi.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinoue T., Weisenberger D.J., Lange C.P., Shen H., Byun H.-M., Berg D.V.D., Malik S., Pan F., Noushmehr H., Van Dijk C.M., et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2011;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kibriya M., Raza A.M., Jasmine F., Roy S., Paul-Brutus R., Rahaman R., Dodsworth C., Rakibuz-Zaman M., Kamal M., Ahsan H. A genome-wide DNA methylation study in colorectal carcinoma. BMC Med. Genom. 2011;4:50. doi: 10.1186/1755-8794-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naumov V.A., Generozov E.V., Zaharjevskaya N.B., Matushkina D.S., Larin A.K., Chernyshov S.V., Alekseev M., Shelygin Y., Govorun V.M. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics. 2013;8:921–934. doi: 10.4161/epi.25577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molnár B., Galamb O., Peterfia B., Wichmann B., Csabai I., Bodor A., Kalmár A., Szigeti K.A., Barták B.K., Nagy Z.B., et al. Gene promoter and exon DNA methylation changes in colon cancer development—mRNA expression and tumor mutation alterations. BMC Cancer. 2018;18:695. doi: 10.1186/s12885-018-4609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori T. Epigenetic Up-regulation of C-C Chemokine Receptor 7 and C-X-C Chemokine Receptor 4 Expression in Melanoma Cells. Cancer Res. 2005;65:1800–1807. doi: 10.1158/0008-5472.CAN-04-3531. [DOI] [PubMed] [Google Scholar]
- 48.Yadav S.S., Prasad S.B., Das M., Kumari S., Pandey L.K., Singh S., Pradhan S., Narayan G. Epigenetic Silencing of CXCR4 Promotes Loss of Cell Adhesion in Cervical Cancer. BioMed Res. Int. 2014;2014:581403. doi: 10.1155/2014/581403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Przybylski M., Kozlowska A.K., Pietkiewicz P., Lutkowska A., Lianeri M., Jagodziński P.P. Increased CXCR4 expression in AsPC1 pancreatic carcinoma cells with RNA interference-mediated knockdown of DNMT1 and DNMT3B. Biomed. Pharmacother. 2010;64:254–258. doi: 10.1016/j.biopha.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Guo J.U., Su Y., Zhong C., Ming G.-L., Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song C.-X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.-H., Zhang W., Jian X., et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2010;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W., Zhang X., Lu X., You L., Song Y., Luo Z., Zhang J., Nie J., Zheng W., Xu D., et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Z., Laird P.W. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenbloom K.R., Sloan C.A., Malladi V., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G., et al. ENCODE data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res. 2012;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen J., El-Gebali S., Natoli M., Sengstag T., Delorenzi M., Bentz S., Bouzourene H., Rumbo M., Felsani A., Siissalo S., et al. Defining new criteria for selection of cell-based intestinal models using publicly available databases. BMC Genom. 2012;13:274. doi: 10.1186/1471-2164-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin I.-H., Chen Y.-F., Hsu M.-T. Correlated 5-Hydroxymethylcytosine (5hmC) and Gene Expression Profiles Underpin Gene and Organ-Specific Epigenetic Regulation in Adult Mouse Brain and Liver. PLoS ONE. 2017;12:0170779. doi: 10.1371/journal.pone.0170779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H., D’Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madrid A., Chopra P., Alisch R. Species-Specific 5 mC and 5 hmC Genomic Landscapes Indicate Epigenetic Contribution to Human Brain Evolution. Front. Mol. Neurosci. 2018;11:39. doi: 10.3389/fnmol.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai J., Chen L., Zhang Z., Zhang X., Lu X., Liu W., Shi G., Ge Y., Gao P., Yang Y., et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195–2205. doi: 10.1136/gutjnl-2019-318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandberg T., Stuart M.P.M.E., Oosting J., Tollenaar R., Sier C.F.M., Mesker W.E. Increased expression of cancer-associated fibroblast markers at the invasive front and its association with tumor-stroma ratio in colorectal cancer. BMC Cancer. 2019;19:284. doi: 10.1186/s12885-019-5462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandberg T., Oosting J., Van Pelt G.W., Mesker W.E., Tollenaar R.A.E.M., Morreau H. Molecular profiling of colorectal tumors stratified by the histological tumor-stroma ratio—Increased expression of galectin-1 in tumors with high stromal content. Oncotarget. 2018;9:31502–31515. doi: 10.18632/oncotarget.25845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibata D., Peinado M.A., Lonov Y., Malkhosyan S., Perucho M. Genomic instability in repeated sequences is an early somatic event in colorectal tumorigenesis that persists after transformation. Nat. Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 63.Aaltonen L.A., Peltomaki P., Mecklin J.P., Järvinen H., Jass J.R., Green J.S., Lynch H.T., Watson P., Tallqvist G., Juhola M. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res. 1994;54:54. [PubMed] [Google Scholar]
- 64.Slattery M.L., Pellatt D.F., Mullany L.E., Wolff R.K., Herrick J.S. Gene expression in colon cancer: A focus on tumor site and molecular phenotype. Genes, Chromosom. Cancer. 2015;54:527–541. doi: 10.1002/gcc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spainhour J.C., Lim H.S., Yi S.V., Qiu P. Correlation Patterns Between DNA Methylation and Gene Expression in The Cancer Genome Atlas. Cancer Informatics. 2019;18 doi: 10.1177/1176935119828776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cappelli E., Felici G., Weitschek E. Combining DNA methylation and RNA sequencing data of cancer for supervised knowledge extraction. BioData Min. 2018;11:22. doi: 10.1186/s13040-018-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Issa J.-P.J., Vertino P.M., Wu J., Sazawal S., Celano P., Nelkin B., Hamilton S.R., Baylin S.B. Increased Cytosine DNA-Methyltransferase Activity During Colon Cancer Progression. J. Natl. Cancer Inst. 1993;85:1235–1240. doi: 10.1093/jnci/85.15.1235. [DOI] [PubMed] [Google Scholar]
- 68.Ramos E.A.S., Grochoski M., Braun-Prado K., Seniski G.G., Cavalli I.J., Ribeiro E.M.S.F., Camargo A.A., Costa F.F., Klassen G. Epigenetic Changes of CXCR4 and Its Ligand CXCL12 as Prognostic Factors for Sporadic Breast Cancer. PLoS ONE. 2011;6:e29461. doi: 10.1371/journal.pone.0029461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mustafi R., Dougherty U., Mustafi D., Ayaloglu-Butun F., Fletcher M., Adhikari S., Sadiq F., Meckel K., Haider H.I., Khalil A., et al. ADAM17 is a Tumor Promoter and Therapeutic Target in Western Diet-associated Colon Cancer. Clin. Cancer Res. 2016;23:549–561. doi: 10.1158/1078-0432.CCR-15-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seidelin J., Horn T., Nielsen O.H. Simple and efficient method for isolation and cultivation of endoscopically obtained human colonocytes. Am. J. Physiol. Liver Physiol. 2003;285:G1122–G1128. doi: 10.1152/ajpgi.00533.2002. [DOI] [PubMed] [Google Scholar]
- 71.Khalil H., Nie W., Edwards R.A., Yoo J. Isolation of Primary Myofibroblasts from Mouse and Human Colon Tissue. J. Vis. Exp. 2013 doi: 10.3791/50611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khare S., Wilson D.M., Tien X.Y., Dudeja P.K., Wali R.K., Sitrin M.D., A Brasitus T. 1,25-Dihydroxycholecalciferol rapidly activates rat colonic particulate guanylate cyclase via a protein kinase C-dependent mechanism. Endocrinology. 1993;133:2213–2219. doi: 10.1210/endo.133.5.8104780. [DOI] [PubMed] [Google Scholar]
- 73.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 74.Li L.-C., Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 75.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Współczesna Onkol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seaborn: Statistical Data Visualization-Seaborn 0.9.0 Documentation, Seaborn.Pydata.org. [(accessed on 8 September 2019)];2018 Available online: http://seaborn.pydata.org/
- 77.Krook M.A., Hawkins A.G., Patel R.M., Lucas D.R., Van Noord R., Chugh R., Lawlor E.R. A bivalent promoter contributes to stress-induced plasticity of CXCR4 in Ewing sarcoma. Oncotarget. 2016;7:61775–61788. doi: 10.18632/oncotarget.11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wegner S.A., Ehrenberg P.K., Chang G., Dayhoff D.E., Sleeker A.L., Michael N.L. Genomic organization and functional characterization of the chemokine receptor CXCR4, a major entry co-receptor for human immunodeficiency virus type 1. J. Biol. Chem. 1998;273:4754–4760. doi: 10.1074/jbc.273.8.4754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.