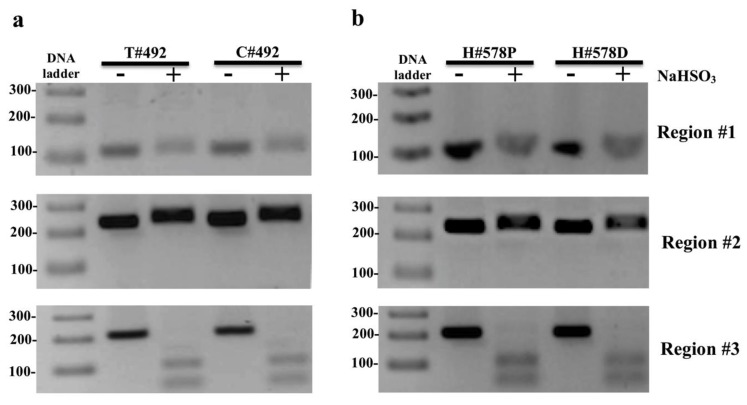
Figure 2.
Representational combined bisulfite restriction analysis (COBRA) results of 3 regions of interest located in CXCR4 from one colorectal cancer (CRC) patient: (a) (#492) and one healthy individual (Supplementary Figure S3) (b) (#578) (Supplementary Figure S4). Each sample was run on two lanes: lane 1) bisulfite-treated PCR amplified DNA without restriction enzyme digestion, which served as a reference control for unmethylated CpG and lane 2) bisulfite-treated PCR amplified DNA digested with a restriction enzyme recognizing amplicons containing a 5’mCpG sequence. Region #3 shows complete PCR product digestion, indicating that region #3 possesses cytosine methylation (5mC) CXCR4 in tumor colonocytes (T# patient ID number), adjacent normal colonocytes (C# patient ID number), and proximal/distal colon of healthy individuals (H# patient ID number P/D proximal/distal colon). Restriction enzyme digestion was not evident in regions #1 or #2 in any tumor or normal colonocytes from any CRC patient or controls and therefore unmethylated in both regions. PCR product treated with restriction enzyme lane = +; PCR product from untreated sample lane = −; Restriction enzyme sensitive digestion was not detected for the amplicons of the intragenic CpG island region #2 or promoter region #1 in either tumor or control samples, indicating the absence of 5mC in those regions (Figure 2a). In addition to CRC samples, regions #1–#3 were assessed in colonocytes from proximal and distal colon in five healthy control individuals. The 5mC results of CXCR4 for colonocytes from healthy control individuals were identical to those of malignant colonocytes from CRC patients, with 5mC present in region #3, but absent in regions #1 or #2 (Figure 2b). Whole COBRA gel images of region #1–#3 for the other CRC and normal control colons tested for CXCR4 5mC are shown in Supplementary Figures S3 and S4, respectively.