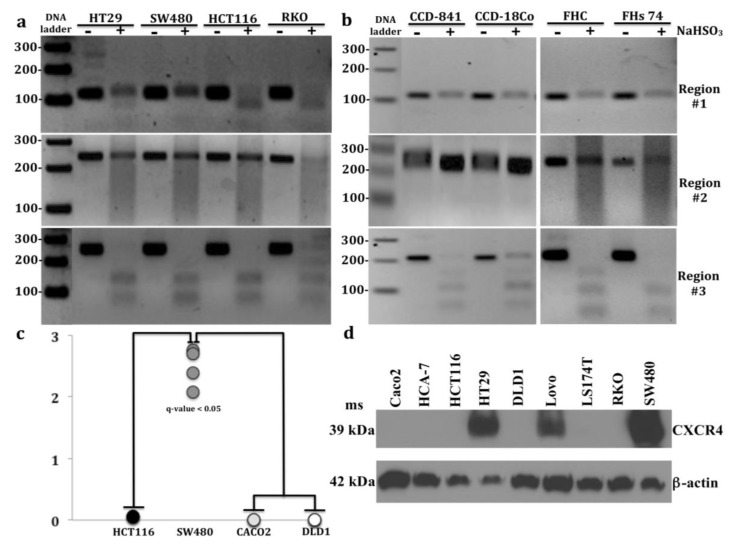
Figure 4.
Combined bisulfite restriction analysis (COBRA) results of 3 regions of interest located in CXCR4 from: (a) 4 CRC cell lines: HT29, SW480, HCT116, and RKO cells and (b) 4 untransformed colon cell lines: CCD-841, CCD-18Co, FHC, and FHs 74 cells. Region #3 shows complete PCR product digestion in CRC cell lines: HT29, SW480, and HCT116 cells and untransformed cell lines: FHC and FHs 74 cells, indicating that region #3 is fully methylated. Incomplete PCR product digestion in RKO cells and untransformed colon cell lines: CCD-841 and CCD-18Co cells indicate partial methylation in region #1. Incomplete PCR digestion was only evident in RKO cells in region #2 indicating partial methylation. 3 of 4 CRC cell lines: HT29, HCT116, and RKO cells displayed incomplete PCR digestion in region #1, whereas no PCR digestion was evident in any untransformed colon cell line cells. (c) RT-PCR analysis of CXCR4 transcript abundance in 4 CRC cell lines: HCT116, SW480, Caco2, and DLD1 cells (q-value < 0.05) after multiple testing correction (Tukey post hoc test, Supplementary Table S1). Note the increase of CXCR4 transcripts in SW480 cells compared to HCT116, Caco2, and DLD1 cells. (d) Western blot analysis of CXCR4 in 9 CRC cell lines: Caco2, HCA-7, HCT116, HT29, DLD1, Lovo, LS174T, RKO, and SW480 cells (Supplementary Table S2). PCR product treated with restriction enzyme lane = +; PCR product untreated lane = −.