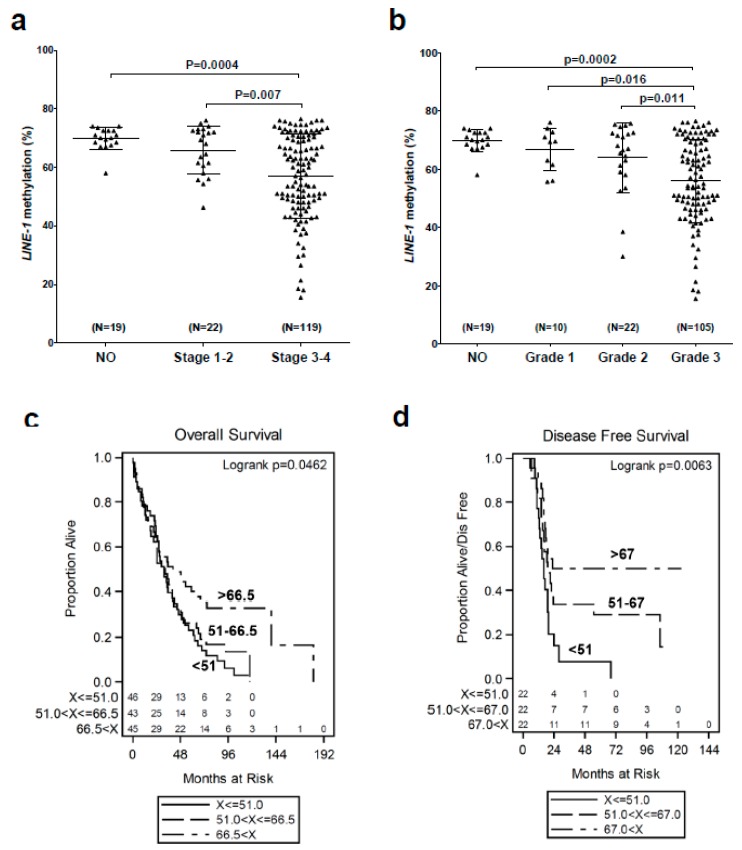
Figure 1.
LINE-1 hypomethylation is associated with advanced disease and reduced survival in EOC. LINE-1 methylation was determined by sodium bisulfite pyrosequencing. (a) LINE-1 methylation vs. clinical stage. (b) LINE-1 methylation vs. pathological grade. For a and b, mean ± SD is plotted, and Mann–Whitney p-values are indicated. (c,d) Kaplan–Meier survival analyses and log rank test p-values of EOC patients separated based on tumor LINE-1 methylation values. (c) LINE-1 methylation vs. overall survival. Patients were separated into three groups based on LINE-1 methylation values: low (<51.0%), middle (51.0–66.5%), and high (>66.5%). A key for the three groups, and the number of patients in each, is shown. (d) LINE-1 methylation vs. disease-free survival. Patients were separated into three groups based on LINE-1 methylation values: low (<51.0%), middle (51.0–67.0%), and high (>67.0%). A key for the three groups, and the number of patients in each, is shown.