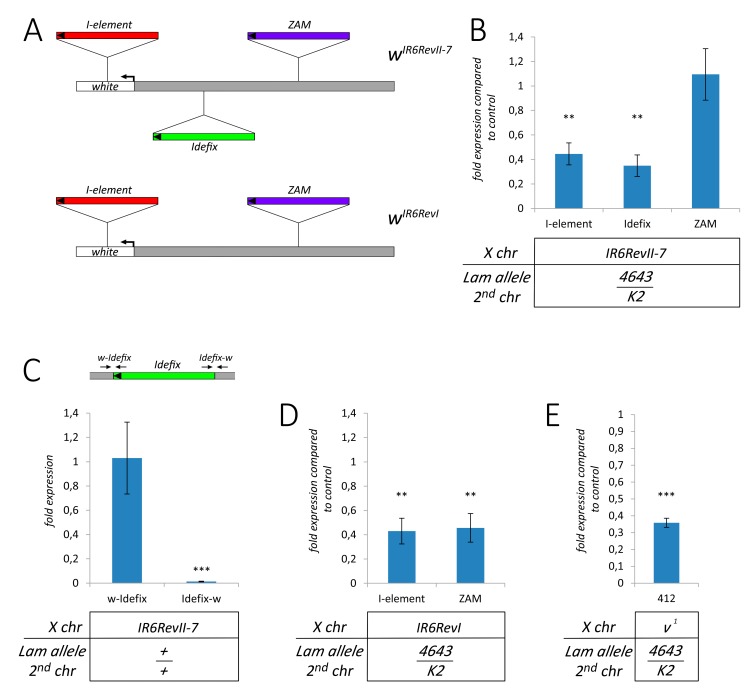
Figure 3.
Silencing of euchromatic retrotransposons in somatic tissues of Lam mutant flies. (A) Schematic representation of the structure of the wIR6RevII7 and wIR6RevI alleles. The I-element (5.4 kb in length) is located in the first intron of the w gene. The Idefix element (7.4 kb in length) is located at about 1660 bp from the w transcription start site. The ZAM element (9 kb in length) is located at about 3 kb from the w transcription start site in the wIR6RevI allele, and more than 10 kb in the wIR6RevII7 alleles. Transcription orientation is indicated by arrows. (B–E) qRT-PCR analysis of some specific retrotransposons in RNAs isolated from female head tissues. Data are mean values from three independent experiments, and error bars indicate SD (** P < 0.01; *** P < 0.005). (B) I-element, Idefix, and ZAM expression in Lam4643/LamK2 mutants (4643/K2) compared to control flies (+/+) in the wIR6RevII7 mutant background. (C) Upper part: schematic representation of the genomic region containing the 5′ upper region of the white gene (grey) where the Idefix element (green) is inserted. The two qPCR couples of primers are represented by black arrows (see Table S1). Lower part: expression of the regions containing the boundaries between Idefix and the white 5′ regions in wIR6RevII7 mutants. w-Idefix is the amplicon containing the boundary between the white 5′ untranscribed regions and Idefix, while Idefix-w is the amplicon containing the boundary between Idefix and the genomic region that separates Idefix from ZAM. (D) I-element and ZAM expression in Lam4643/K2 mutants (4643/K2) compared to control flies (+/+) in the wIR6RevI mutant background. (E) Expression of the 412 retrotransposon in Lam04643/K2 mutants compared to the Lam+/+ in the v1 mutant background.