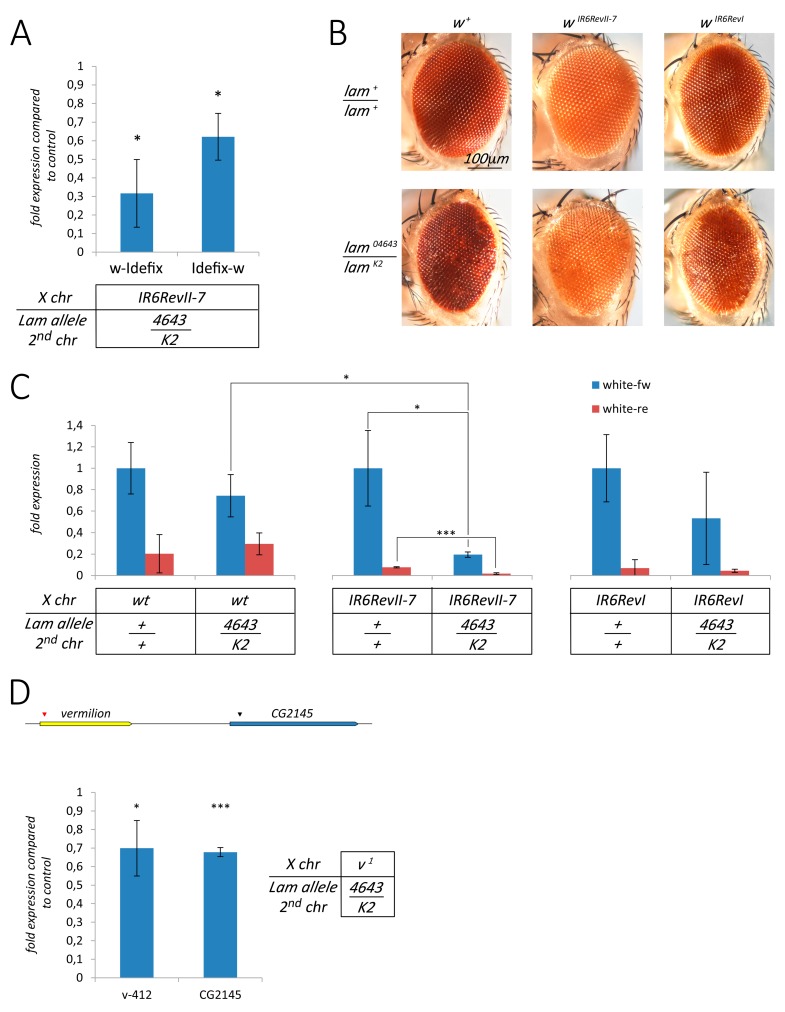
Figure 5.
Silencing of the genes located in the neighborhood of silenced euchromatic retrotransposons in Lam mutant flies. (A) qRT-PCR analysis of the regions containing the boundaries between Idefix and the white untranscribed regions of wIR6RevII7; Lam04643/K2 compared to wIR6RevII7; Lam+. Data are mean values from three independent experiments, and error bars indicate SD (* P < 0.05). (B) Representative bright-field microscope images of adult eyes of wild type, wIR6RevII7, and wIR6RevI flies with and without lam loss-of-function mutant allelic combination. (C) qRT-PCR analysis of white expression in RNAs isolated from female head tissues. Blue histograms represent sense expression, while red histograms represent anti-sense expression. white expression in each genetic background is referred to the sense expression of its specific control. Data are mean values from three independent experiments, and error bars indicate SD (* P < 0.05; *** P < 0.001). (D) Upper part: schematic representation of the genomic region containing the vermilion and CG2145 genes. Black arrowhead indicates the 412 insertion point where v-412 primers were designed, while red arrowhead indicates the region where CG2145 primers were designed (see Table S1). Lower part: qRT-PCR analysis in RNAs isolated from female head tissues in Lam04643/K2 mutants compared to the Lam+/+ in the v1mutant background. Data are mean values from three independent experiments, and error bars indicate SD (* P < 0.05; *** P < 0.005).