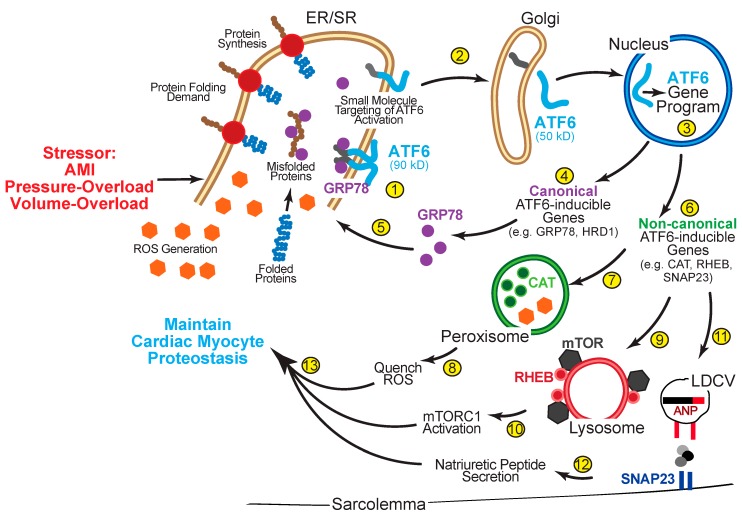
Figure 2.
ATF6 activation and gene program induction. ① In its inactive state, ATF6 is a 90kD ER transmembrane protein that is anchored in the membrane in oligomers via GRP78 and intermolecular disulfide bonding. ② Upon stressors, like cardiac hypertrophy that increases protein synthesis and the protein folding demand or AMI that elevates cellular levels of reactive oxygen species (ROS), GRP78 dissociates from the ER luminal domain of ATF6 and the disulfide bonds are reduced allowing monomerization of ATF6, which allows the 90 kD form of ATF6 to translocate to the Golgi, where is it cleaved by S1P and S2P to liberate the N-terminal approximately 400 amino acids (50 kD) of ATF6 from the ER membrane. It is this unique sequence of activation steps that open a window of small molecule targeting and discovery of ATF6-based therapeutics. ③ The clipped form of ATF6 has a nuclear localization sequence, which facilitates its movement to the nucleus where it binds to specific regulatory elements in ATF6-responsive genes, such as ER stress response elements (ERSEs), and induces the ATF6 gene program. ④ The canonical ATF6 gene program comprises genes that encode proteins that localize to the ER, such as the chaperone, GRP78, or components of ERAD, HRD1 ⑤, where they fortify ER protein folding. ⑥ The non-canonical ATF6 gene program comprises genes that encode proteins not typically categorized as ER stress-response proteins, many of which localize to regions of the cell outside the ER. ⑦ Catalase is a potent antioxidant that localizes to the lumen of peroxisomes where it functions to ⑧ quench ROS. ⑨ Rheb is a small GTPase located on the surface of lysosomes that when bound to mTOR, ⑩ promotes mTORC1 activation and sustains protein synthesis. ⑪ SNAP23 is a t-SNARE protein crucial for ⑫ secretion of natriuretic peptides via large dense-core vesicles (LDCV) in response to hemodynamic stress. ⑬ Both the canonical and non-canonical ATF6 gene programs coordinate to maintain cardiac myocyte proteostasis.