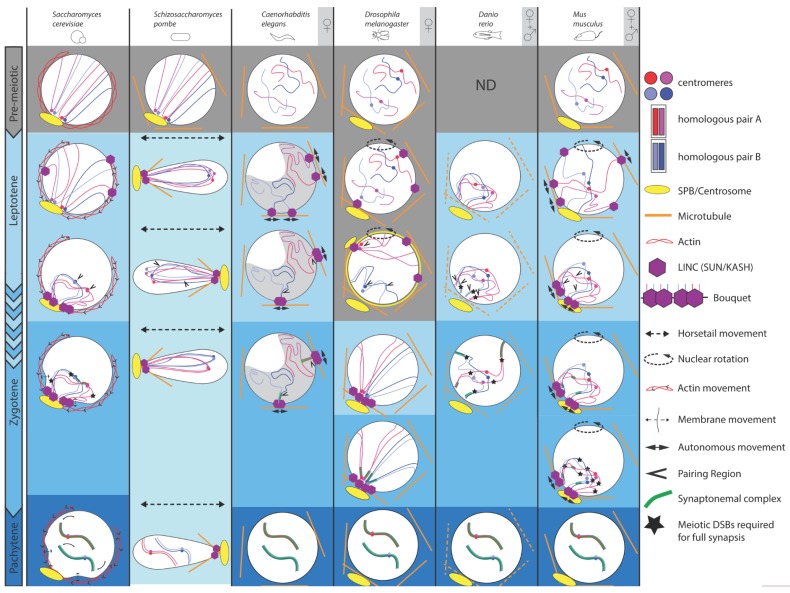
Figure 1.
Comparison of chromosomal and nuclear meiotic movements in different species. Yeasts, C. elegans, Drosophila, zebrafish and mice exhibit dramatic changes throughout meiotic prophase I substages, i.e., leptotene, zygotene, pachytene and diakinesis (not shown). These substages are well defined, except for S. pombe, which has no synaptonemal complex (SC; lightest blue), but displays peculiar LINE structures reminiscent of SC. For our selected sexually reproducing models, data were obtained either only in females ♀ (oogenesis), or also from males (♀ + ♂) (spermatogenesis). At pre-meiotic stages (grey background) in yeasts and mouse, Rabl orientation is inherited from previous divisions, when centromeres (blue and red dots) are pulled toward the spindle pole body or centrosome (yellow oval with emanating microtubules), with trailing telomeres. C. elegans, Drosophila and mouse do not show such organization. Zebrafish chromosomes organization has not been defined at this stage. Chromosome dynamics start from early leptotene, except in Drosophila, where movements take place during premeiotic divisions (grey background). Nuclear movements (dashed arrows) occur in yeasts, Drosophila and mouse, thus generating coordinated chromosome dynamics, whereas solitary chromosome movements (plain arrows) occur in C. elegans and, in addition to coordinated movements, in mouse. These movements rely on the coordinated action of the microtubule cytoskeleton (except in S. cerevisiae, actin), the dynein motor and the LINC complex (SUN/KASH). In S. cerevisiae, early movements depend on actin, whose rapid polymerization cycles push on the nucleus, consequently shaking chromosomes. At pachytene, telomeres display abrupt movements concomitant with nuclear membrane deformations. In S. pombe, prophase I-like stage is called the horsetail stage: the nucleus elongates and moves back and forth between the ends of the cell. Telomeres remain clustered at the leading edge of the moving nucleus. Drosophila and mouse display entire nuclear rotations driven by the microtubule cytoskeleton. Solitary microtubules-driven movements (plain arrows) have also been identified in C. elegans through the coordinated action of microtubules, dynein and LINC complex at the nuclear membrane. Concomitantly with early movements, centromeres move away from the pole, while telomeres attach to the nuclear membrane and move to a small area adjacent to the spindle pole, forming a bouquet at leptotene/zygotene transition in yeast and mouse, or early leptotene in zebrafish. C. elegans oocytes pack their chromatin in a characteristic half-moon territory. Resulting proximity of specific chromosomal regions (see text) leads to initial homologous pairing (arrowheads). Except for zebrafish, initial pairing is DSB-independent. Entry in zygotene is marked by the initiation of the synaptonemal complex formation (green ladder-like structure). During zygotene, chromosomes move out of the bouquet. Completion of synapsis and resolution of interlocks marks pachytene, displaying well-separated chromosome pairs. Full synapsis of homologues requires DSBs, except in C.elegans and Drosophila.