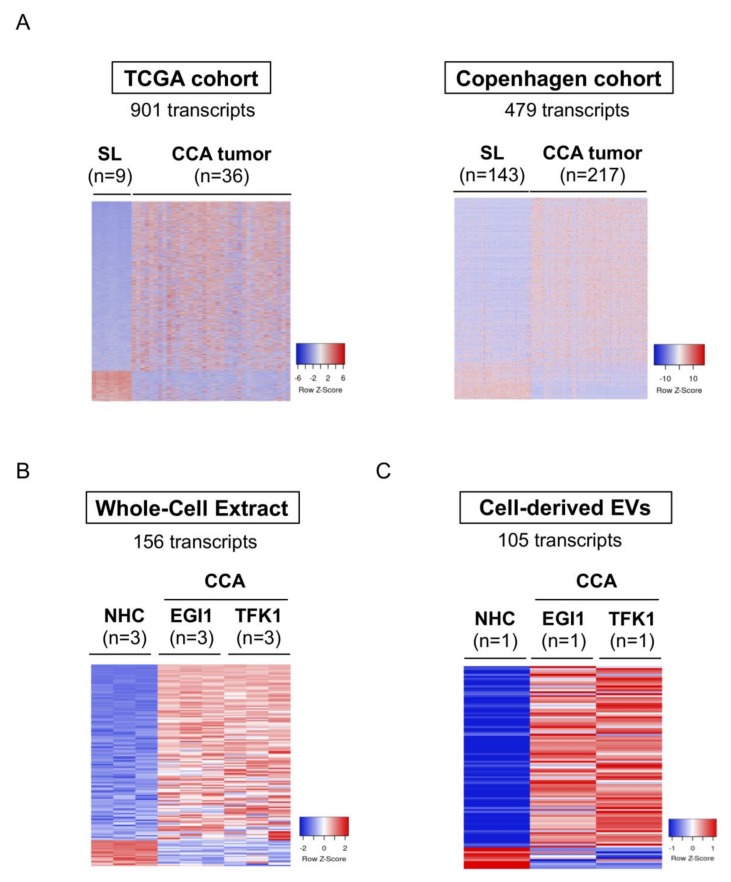
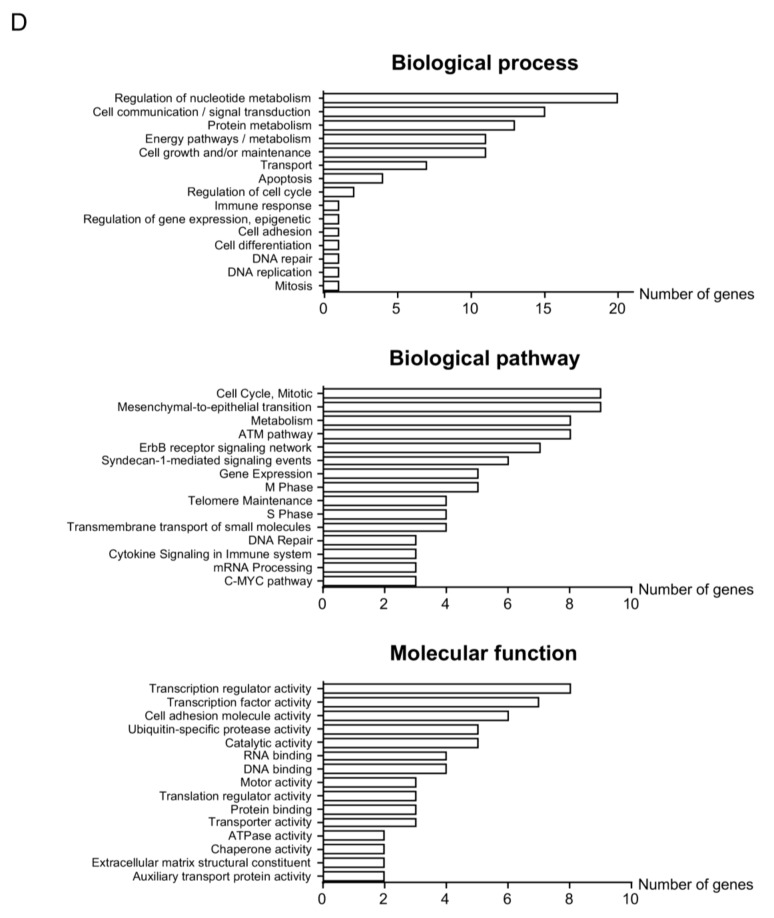
Figure 4.
mRNAs commonly deregulated between serum EVs, CCA tumors from two independent cohorts of patients, tumor cells in vitro and in CCA-derived EVs. mRNAs differentially abundant in serum EVs from patients with CCA vs. (PSC + UC + Healthy individuals) were compared with the transcriptome of: (i) patients with CCA from TCGA (n = 36) and “Copenhagen” (n = 217) cohorts, (ii) CCA cells (EGI1 and TFK1) and cell-derived EVs compared to their respective control groups, further selecting the ones that are commonly expressed. Heatmap of the differentially expressed transcripts in (A) TCGA (left) and “Copenhagen” cohorts (right); (B) Whole-cell extracts from CCA cells and NHCs; (C) Cell-derived EVs; (D) Gene ontology (GO: FunRich database [27]) analysis of the 105 transcripts commonly altered in serum EVs, CCA human tumors, CCA cells and in cell-derived EVs, highlighting the biological processes and pathways in which the identified transcripts are involved, as well as their biological function. Abbreviations: CCA, cholangiocarcinoma, EVs, extracellular vesicles; NHCs, normal human cholangiocytes; SL, surrounding liver; TCGA, The cancer genome atlas.