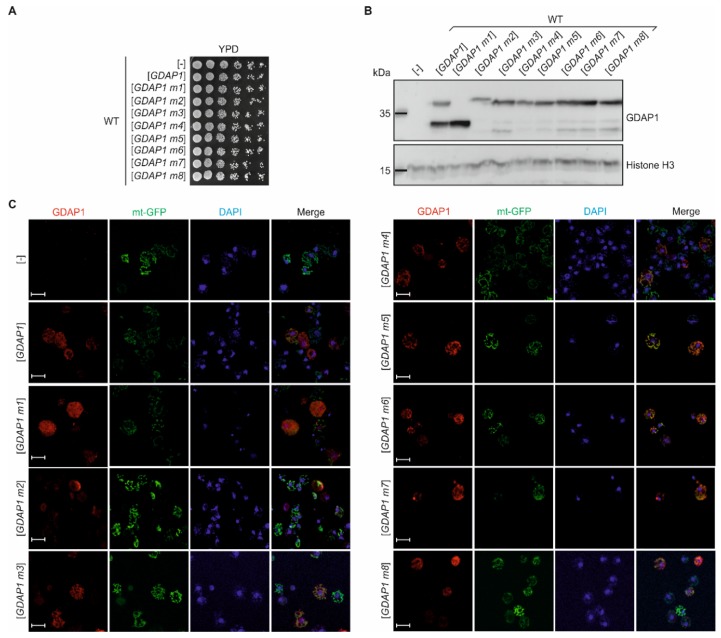
Figure 2.
Yeast-based system to analyze GDAP1 protein function. (A) Overnight cultures of wild-type yeast cells harboring empty vector ([-]) or plasmids with GDAP1 gene variants were diluted to OD600 ≈ 1 and then ten-fold serial dilutions were prepared and spotted on to YPD medium. Plates were incubated for 2 d at 28 °C. (B) Yeast wild-type cells harboring empty vector ([-]) or plasmids with GDAP1 gene variants (as indicated) were cultured overnight in SC-leu at 28 °C, then were collected and disrupted. The total cell extracts obtained were analyzed by SDS-PAGE, followed by immunoblotting with anti-GDAP1 or anti-Histone H3 antibodies, as indicated. (C) Yeast wild-type cells transformed with plasmid encoding mitochondrially targeted green fluorescent protein (mt-GFP) and with empty vector ([-]) or plasmids encoding GDAP1 alleles (as indicated) were grown in SC–leu-his medium overnight and then were shifted to glycerol-containing medium for 4 h to induce expansion of mitochondria. Cells were fixed, stained and observed using confocal microscopy. GDAP1 was visualized by immunostaining (red), mitochondria using the mt-GFP protein marker (green) and the nucleus was stained with DAPI (blue). Scale bar 5 µm.