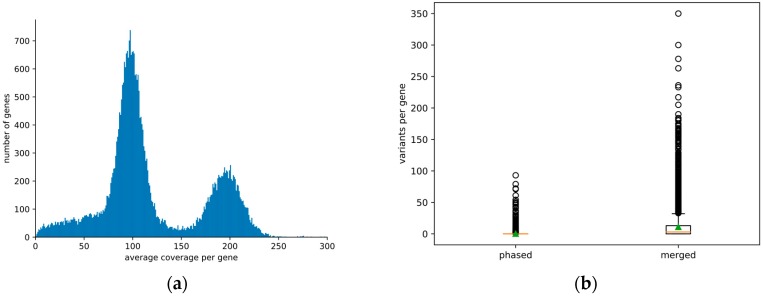
Figure 1.
(a) Distribution of the average sequencing read depth per gene model. Predicted gene models were classified into phase-separated and merged based on the average read depth value deduced from the analysis presented here. The haploid read depth with Illumina short reads ranges from 50-fold to 150-fold. (b) Number of heterozygous sequence variants in phase separated and merged genes. The high proportion of heterozygous variants in merged gene models is due to the mapping of reads originating from two different alleles to the same region of the assembly.