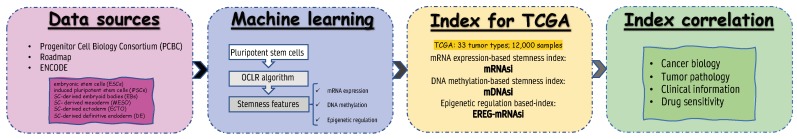
Figure 1.
The overall methodology of the Stemness Indexes development [22]. The data sources from the Progenitor Cell Biology Consortium (https://www.synapse.org/pcbc), Roadmap (http://www.roadmapepigenomics.org), and the ENCyclopedia Of DNA Elements (ENCODE) Project (https://www.genome.gov/Funded-Programs-Projects/ENCODE-Project-ENCyclopedia-Of-DNA-Elements) were profiled using the Illumina HumanMethylation 450 (HM450) platform to define stem cell signatures. The pluripotent stem cells include four embryonic stem cells (ESCs), 40 induced pluripotent stem cells (iPSCs), 22 stem cell (SC)-derived embryoid bodies (EBs), 11 SC-derived mesoderm (MESO), 11 SC-derived ectoderm (ECTO), and 11 SC-derived definitive endoderm (DE). The one-class logistic regression (OCLR) algorithm was used to train for stemness features, on stem cell (SC; ESC/iPSC) classes and their differentiated progenitors. As a result, the mRNA expression-based stemness index (mRNAsi), DNA methylation-based stemness index (mDNAsi), and epigenetic regulation based-index (EREG-mRNAsi) were obtained. These stemness indices have also been applied to datasets from TCGA in order to calculate the mRNAsi, EREG-mRNAsi, and mDNAsi scores of the samples. The indexes for each TCGA cases were validated by the correlations with known cancer biology, tumor pathology, clinical information, and drug resistance data.