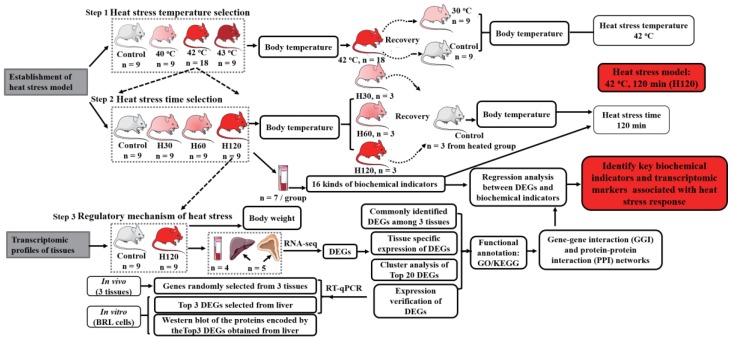
Figure 1.
Experimental design of the study. First, heat stress (HS) conditions were explored by analyzing the body temperature of rats at different high ambient temperatures or treatment times, as well as body temperature recovery after transferring to 30 °C or 22 ± 1 °C from HS conditions (Steps 1 and 2). After 42 °C HS for 30 min (H30), 60 min (H60), and 120 min (H120), animals were euthanized, and their blood biochemical indicators were measured (Step 2). RNA-sequencing was used to establish the transcriptomic profiles of blood (n = 4), liver (n = 5), and adrenal gland (n = 5) tissues of rats in H120 and control groups (Step 3). Differentially expressed genes (DEGs) analysis, its validation, gene ontology terms (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and gene–gene and protein–protein interaction network (GGI and PPI) construction were conducted to characterize the common and specific DEGs responses to HS in blood, liver, and adrenal glands. Finally, the Pearson correlation analysis of the DEGs in three tissues and the biochemical indicators in blood was performed.