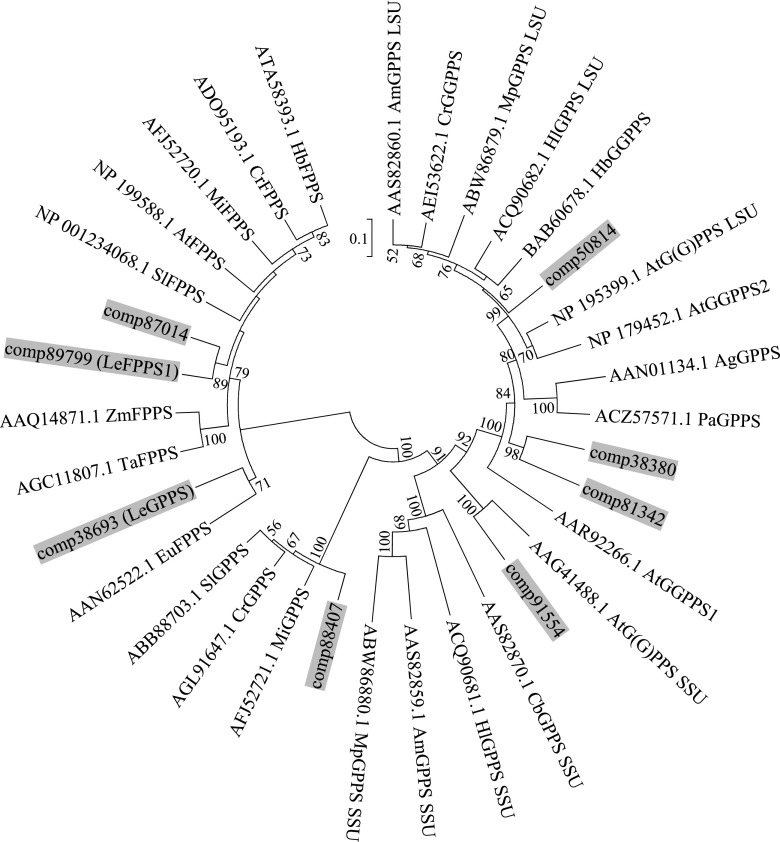
Figure 5.
Unrooted phylogenetic analysis of the deduced amino acid sequences of trans-prenyl diphosphate synthase homologs in L. erythrorhizon and other plant species. The prenyl diphosphate synthases of L. erythrorhizon are highlighted in gray. Three partial sequences of trans-prenyl diphosphate synthase homologs found in RNA-seq data of L. erythrorhizon were not included. The tree was inferred by the bootstrap neighbor-joining method using the program MEGA7 (http://evolgen.biol.se.tmu.ac.jp/MEGA/) with default settings. The percentage of bootstraps, calculated by 1000 repeats, is shown next to each branch, with bootstrap values less than 50% omitted. The GenBank accession numbers are shown for each enzyme. The first two letters indicate the scientific names of each species: At, Arabidopsis; Ag, Abies grandis; Am, Antirrhinum majus; Eu, Eucommia ulmoides; Cb, Clarkia breweri; Cr, Catharanthus roseus; Hb, Hevea brasiliensis; Hl, Humulus luplus; Mi, Mangifera indica; Mp, Mentha x piperita; Pa, Picea abies; Sl, Solanum lycopersicum; Ta, Triticum aestivum; Zm, Zea mays.