Figure 1.
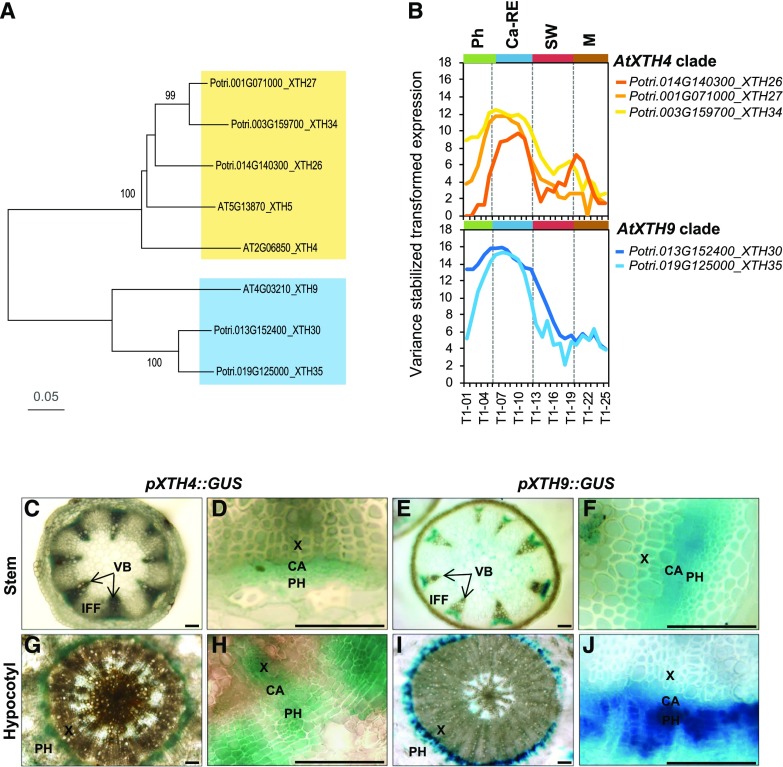
Clades AtXTH4 and AtXTH9 in Arabidopsis and Populus trichocarpa. A, Phylogenic tree constructed using the neighbor-joining method of MEGA7 in default mode using MUSCLE-aligned protein sequences (http://phylogeny.lirmm.fr/phylo_cgi/index.cgi), with bootstrap test of 1,000 replicates shown in percentages beside branches. B, The expression patterns of P. trichocarpa members of AtXTH4 and AtXTH9 clades in different wood developmental zones (http://aspwood.popgenie.org). Ca-RE, Cambium-radial expansion zone; M, maturation zone; Ph, phloem; SW, secondary wall formation zone. C to J, AtXTH4 and AtXTH9 promoter activity in Arabidopsis mature inflorescence stems and hypocotyls as visualized by GUS histochemistry. In the inflorescence stems (C–F), the expression of both genes was detected in vascular bundles, whereas interfascicular fibers did not show any expression (C and E); the closeup vascular bundles (D and F) show signals in the vascular cambium, developing xylem, and developing and differentiated phloem. In hypocotyls (G–J), both genes were expressed in the region of secondary vascular tissue formation (G and I), encompassing the vascular cambium, developing secondary xylem and phloem, and recently differentiated phloem cells (H and J). CA, Vascular cambium; IFF, interfascicular fibers; PH, phloem; VB, vascular bundle; X, xylem. Bars = 50 µm.